- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
In vitro Glutamylation Inhibition of Ubiquitin Modification and Phosphoribosyl-Ubiquitin Ligation Mediated by Legionella pneumophila Effectors
Published: Vol 10, Iss 21, Nov 5, 2020 DOI: 10.21769/BioProtoc.3811 Views: 3921
Reviewed by: David A. CisnerosJose Antonio Reyes-DariasAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

H2 Production from Methyl Viologen–Dependent Hydrogenase Activity Monitored by Gas Chromatography
Nuttavut Kosem
Dec 5, 2023 1758 Views

Monitoring Protein Stability In Vivo Using an Intein-Based Biosensor
John S. Smetana [...] Christopher W. Lennon
Apr 20, 2025 1575 Views

Endo-1,4-β-D-xylanase Assay Using Azo-Xylan and Variants Thereof
Luca Bombardi [...] Salvatore Fusco
Apr 20, 2025 1916 Views
Abstract
Glutamylation is a posttranslational modification where the amino group of a free glutamate amino acid is conjugated to the carboxyl group of a glutamate side chain within a target protein. SidJ is a Legionella kinase-like protein that has recently been identified to perform protein polyglutamylation of the Legionella SdeA Phosphoribosyl-Ubiquitin (PR-Ub) ligase to inhibit SdeA’s activity. The attachment of multiple glutamate amino acids to the catalytic glutamate residue of SdeA by SidJ inhibits SdeA’s modification of ubiquitin (Ub) and ligation activity. In this protocol, we will discuss a SidJ non-radioactive, in vitro glutamylation assay using its substrate SdeA. This will also include a second reaction to assay the inhibition of SdeA by using both modification of free Ub and ligation of ADP-ribosylated Ubiquitin (ADPR-Ub) to SdeA’s substrate Rab33b. Prior to the identification and publication of SdeA’s activity, no SdeA inhibition assays existed. Our group and others have demonstrated various methods to display inhibition of SdeA’s activity. The alternatives include measurement of ADP-ribosylation of Ub using radioactive NAD, NAD hydrolysis, and Western blot analysis of HA-Ub ligation by SdeA. This protocol will describe the inhibition of both ubiquitin modification and the PR-Ub ligation by SdeA using inexpensive standard gels and Coomassie staining.
Keywords: SidJBackground
Legionella pneumophila is an infectious bacterium that opportunistically infects alveolar macrophages. This occurs through the inhalation of contaminated water aerosols, causing a potentially lethal form of pneumonia known as Legionnaires’disease (McDade et al., 1977). Legionella infects host cells by the secretion of over 300 effector proteins that are used to hijack many host cellular processes and prevent their lysosomal degradation (Hubber and Roy, 2010). One process hijacked by Legionella is the ubiquitination system (Hubber et al., 2013). Ubiquitination is a eukaryotic posttranslational modification, that regulates a variety of cellular processes (Hershko and Ciechanover, 1998; Chen and Sun, 2009; Hurley and Stenmark, 2011; Haglund and Dikic, 2012). This requires a concerted effort of E1, E2, and E3 enzymes to carefully regulate which proteins are ubiquitinated (Scheffner et al., 1995). However, Legionella have co-opted this modification with the SidE family of phosphoribosyl-ubiquitin ligases that act independently of E1 and E2 enzymes. This family uses two catalytic domains to ADP-ribosylate ubiquitin using the mono-ADP-ribosyl transferase (mART) domain, and then, ligates ADPR-Ub to a host protein serine residues using a phosphodiesterase (PDE) domain (Bhogaraju et al., 2016; Qiu et al., 2016; Kotewicz et al., 2017). These previously mentioned studies include means for assaying SidE activity. The SidE family includes a member, SdeA, which has been identified to be spatiotemporally regulated by SidJ (Havey and Roy, 2015; Jeong et al., 2015; Urbanus et al., 2016); although, the mechanism of this regulation was not completely understood. It was suggested that SidJ may act as a PR-deubiquitinase (Qiu et al., 2017) when using Legionella purified SidJ (Qiu and Luo, 2019); however, recent studies do not recapitulate these results (Bhogaraju et al., 2019; Wan et al., 2019; Shin et al., 2020).
Our group (Sulpizio et al., 2019), and several others (Bhogaraju et al., 2019; Black et al., 2019; Gan et al., 2019), have demonstrated that SidJ is a polyglutamylase that adds multiple glutamate amino acids to SdeA to inhibit SdeA’s function. These studies provided structural data that identified SidJ contains a kinase-like domain and binds the eukaryotic protein calmodulin. Furthermore, mass spectrometry studies have identified that the mechanism of SdeA’s inhibition is polyglutamylation of SdeA’s catalytic mART glutamate residue. Based on these results, an in vitro glutamylation assay was developed using the substrate SdeA, calmodulin, ATP/MgCl2, and glutamate. To demonstrate the inhibitory effect of glutamylation, in vitro glutamylation was followed by a SdeA activity assay. Other groups have also described SdeA inhibition using NAD hydrolysis (Bhogaraju et al., 2019), radioactive NAD (Black et al., 2019), Flag-tagged SdeA substrates (Gan et al., 2019), HA-Ub, and HA-Ub variants resistant to canonical ubiquitination by these groups. These alternative methods are suitable for identifying inhibition and may provide more quantitative detection. However, some of these experiments did not include in vitro inhibited SdeA, and those that monitor ubiquitin modification require the use of expensive reagents such as radioactive NAD. This protocol discusses a glutamylation assay developed for SidJ and assays in vitro inhibition of both SdeA PR-ubiquitin ligation and ubiquitin modification using standard gels and Coomassie staining. This can be used to identify the effects of mutations on activity, assay inhibition of both ubiquitin modification and PR-Ubiquitin ligation, and may more generally be adopted to determine inhibition of other ADP-ribosyl transferases by glutamylation.
Materials and Reagents
1.7 ml Microtubes (Corning Incorporated, Axygen, catalog number: MCT-175-C )
50 ml Centrifuge tubes (VWR, catalog number: 525-0637 )
Gloves (VWR, catalog number: 89038-270 )
Kim wipes (Kimberly-Clark Professional, catalog number: 34120 )
Pipette tips:
10 μl XL Graduated Tips (USA Scientific, Tip One, catalog number: 1110-3700 )
200 μl Graduated Quick Rack (Laboratory Products Sales, catalog number: 130430 )
1,250 μl Pipette tips (Laboratory Product Sales, catalog number: L134770)
Recombinant proteins
SidJ 89-853 truncation, SdeA 211-1152 truncation, human calmodulin 2, were expressed in Escherichia coli with an N-terminal 6XHis-SUMO tag and purified as described previously (Sulpizio et al., 2019). Rab33b (1-200) was also purified as proteins mentioned in (Sulpizio et al., 2019) and Ub was purified as described in Akturk et al. (2018). Final purified proteins were stored in buffer (20 mM Tris pH 7.5, 150 mM NaCl) without glycerol, aliquoted, flash-frozen, and stored at -80 °C.
β-Nicotinamide adenine dinucleotide sodium salt (NAD) (Sigma, catalog number: N0632-1G )
L-Glutamic Acid Monosodium Salt, Monohydrate (USB Corporation, catalog number: 16245 )
2-Mercaptoethanol (Sigma, catalog number: M3148-100ML )
30% Acrylamide/Bis solution 37.5:1 (Bio-Rad, catalog number: 1610158 )
Acetic acid, glacial (J.T. Baker, catalog number: 9508-06)
Adenosine 5’-triphosphate disodium salt hydrate (Sigma, catalog number: A2382-10G )
Ammonium persulfate (APS) (Amresco, catalog number: 0486-100G )
Brilliant Blue R-250 (Fisher, catalog number: BP101-50 )
Bromophenol Blue sodium salt (Fisher, catalog number: BP114-25 )
DL-Dithiothreitol (DTT) (Amresco, catalog number: M109-25g )
Ethanol 200-proof (Koptec, catalog number: V1001 )
Glycerol (Mallinckrodt Chemicals, catalog number: 5092-16 )
Glycine (VWR, catalog number: 0167-5KG )
Magnesium chloride, 6-hydrate (Mallinckrodt Chemicals, catalog number: 5958-04 )
Methanol (Fisher, catalog number: A454SK-4 )
N,N,N’,N’-tetramethylethylene-diamine (TEMED) (Bio-Rad, catalog number: 161-0800 )
Precision Plus Protein All Blue Standards Protein Ladder (Bio-Rad, catalog number: 161-0373 )
Sodium chloride (VWR, catalog number: 0241-10KG )
Sodium dodecyl sulfate (SDS) (VWR Life Sciences, catalog number: 0227-1KG )
Tris (VWR, catalog number: 0497-5KG )
Reaction Buffer (see Recipes)
MgCl2 1 M Solution (see Recipes)
ATP 100 mM pH 7.5 Solution (see Recipes)
Glutamic Acid 1 M Solution (see Recipes)
10x SDS-PAGE Running Buffer (see Recipes)
10x Native-PAGE Running Buffer (see Recipes)
SDS Sample Buffer (see Recipes)
Native Sample Buffer (see Recipes)
Coomassie Stain (see Recipes)
Coomassie Destaining Solution (see Recipes)
12% SDS-PAGE Resolving Gel (see Recipes)
4% SDS-PAGE Stacking Gel (see Recipes)
8% Native PAGE Resolving Gel (see Recipes)
3% Native PAGE Stacking Gel (see Recipes)
Note: Products were stored as suggested by manufacture except where listed.
Equipment
-80 °C freezer (So-Low, model: PV85-21 )
Computer
Dry Bath (Benchmark, model: BSH1001 )
Fixed speed centrifuge (Benchmark, model: C1008-C )
Forceps
Gel electrophoresis apparatus (Bio-Rad, model: Mini-PROTEAN Tetra System )
Gel electrophoresis power supply (Bio-Rad, model: PowerPac Basic )
Gel imager (Bio-Rad, model: Chemidoc MP Imaging System )
Ice bucket
Labcoat (VWR, catalog number: 10141-306 )
Laboratory tape (VWR, catalog number: 89098-062 )
Microwave (Sharp, model: R230KW )
Pipettes (Gilson, model: Pipetman classic P2, P20, P200, P1000, catalog numbers: F144801 , F123600 , F123601 , F123602 )
Rocking shaker (Reliable Scientific, Inc., model: 55D 12 x 16 )
Sheet protectors (Clear file, Archival Plus 5x7 Print, catalog number: 370100B )
Vortex mixer 120V (Corning LSE, model: 6775 )
Procedure
SidJ in vitro Glutamylation Reaction
Review the flowchart of the experimental outline of SidJ glutamylation and SdeA activity assays before beginning the procedure (Figure 1).

Figure 1. Experimental outline for SidJ in vitro Glutamylation and SdeA Inhibition assay. A flow chart of the general experimental steps described in this procedure. The left branch is to assay the inhibition of SdeA ADP-ribosylation or Phospho-Ribosylation of Ub. The right branch is the experiments to assay PR-Ub ligation to substrates.Thaw recombinantly purified SidJ 89-853, SdeA Core (truncation 211-1152), calmodulin, ubiquitin, and optionally, Rab33b (1-200) on ice.
Note: Rab33b 1-200 was used in this assay due to the prominence of a single PR-Ubiquitination band and increased protein stability (Jon Wasilko, Mao Lab, unpublished results). Full-length Rab33b may also be used instead.
Preheat dry bath to 37 °C.
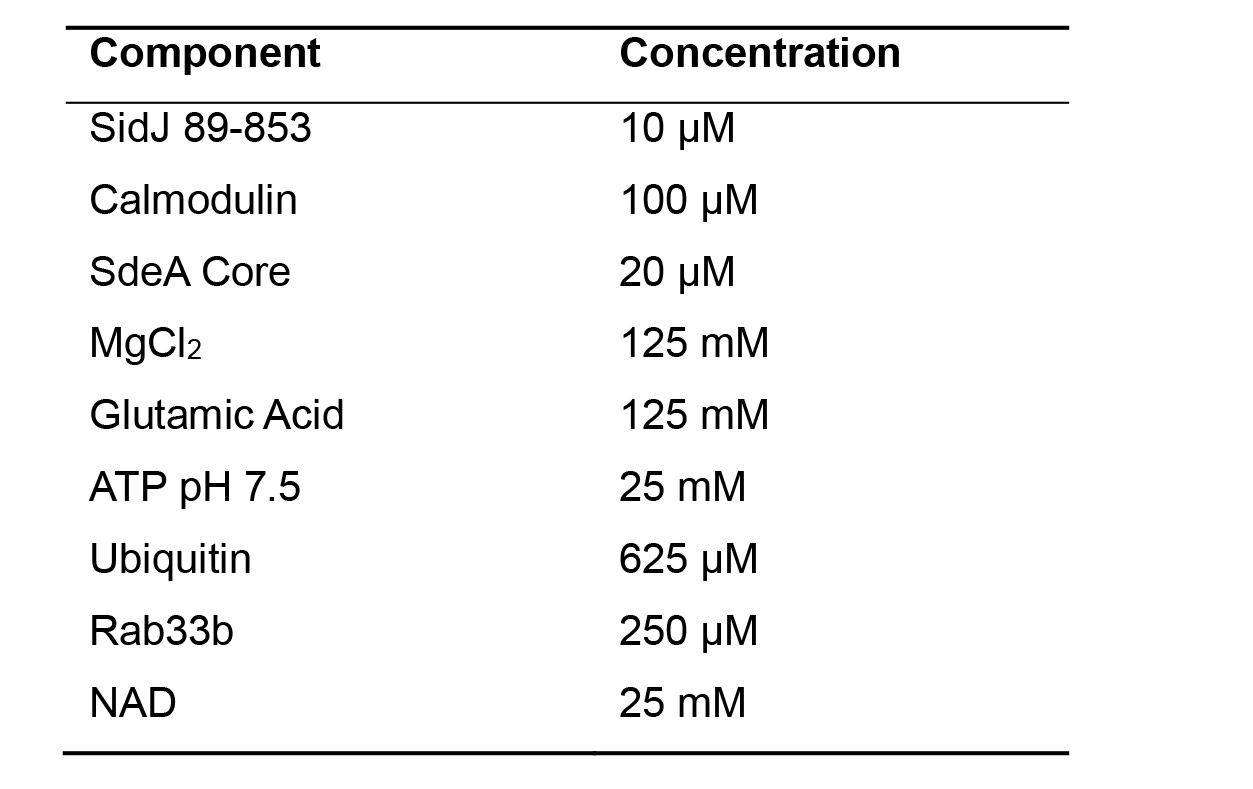
Prepare stock solutions, on ice, listed in Table 1 by diluting in reaction buffer (20 mM Tris pH 7.5, 50 mM NaCl). Prepare NAD solution fresh and use glutamic acid and ATP stocks stored at -80 °C.
Table 1. Stock solution concentrations for SidJ in vitro glutamylation and SdeA inhibition reaction
Pipette the volumes of stock solution listed in SidJ glutamylation reaction row for a 25 μl reaction listed in Table 2 into a chilled 1.7 ml microcentrifuge tube on ice. Pipette ATP last to initiate the reaction. Immediately vortex, centrifuge briefly (~10 s max speed), and incubate the samples at 37 °C in a dry bath for 30 min.
Note: It is important that samples are mixed thoroughly and centrifuged. SdeA is very active and the reaction mixture needs to be homogenous for maximal inhibition.
Optional: Prepare a master mix for Step A5. Combine components contained in all reactions by pipetting stock solutions and 10 μl of reaction buffer per reaction. Prepare approximately 10% more reaction mix than needed for samples. The addition of reaction buffer to the master mix dilutes components to maintain protein stability. If a master mix is prepared, for each reaction, subtract the volumes of reaction components included in the mix and 10 μl of reaction buffer from the amount used in Table 2.
Table 2. SidJ in vitro glutamylation and SdeA modification reaction components and concentrations
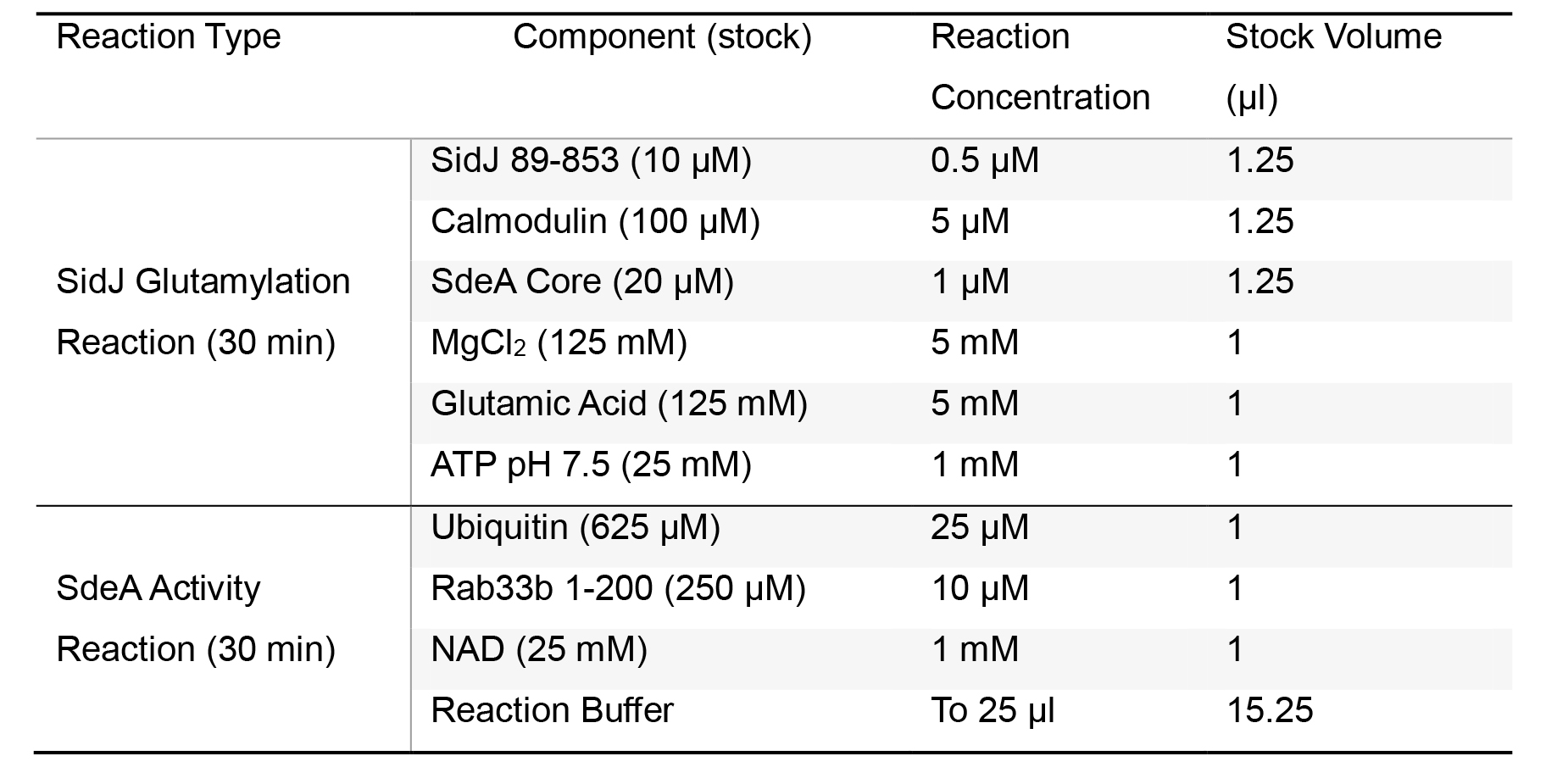
Pipette remaining reaction components for the SdeA activity portion of the assay into the tube. If assaying ubiquitin modification, replace Rab33b with reaction buffer. Immediately vortex, centrifuge briefly (~10 s max speed), and incubate samples at 37 °C in a dry bath for 30 min.
Note: A master mix containing SdeA modification components and 3 μl reaction buffer per sample can be prepared to minimize pipetting inaccuracies. A negative control excluding NAD would be beneficial for the visualization of the absence of SdeA activity.
If assaying ubiquitin modification, separate each reaction into two tubes by pipetting 12.5 μl into another 1.7 ml microcentrifuge tube. Label tubes and pipette 3 μl of native sample buffer into one tube for native-PAGE analysis, and 3 μl of SDS sample buffer into the other for SDS-PAGE analysis and visualization of protein loading. Vortex to mix and centrifuge briefly (10 s at max speed). If only assaying PR-ubiquitin ligation activity, the reaction can be halted by the addition of 6 μl of SDS sample buffer.
Note: Do not include SDS in native sample buffer, native-PAGE gels, or native-PAGE running buffer.
Detection of ubiquitin modification and PR-Ubiquitin ligation to Rab33b by gel electrophoresis
For detection of ubiquitin modification, electrophorese 13 μl of the portion of each sample in native sample buffer using a native-PAGE gel and native-PAGE running buffer at 80 V. Once samples have migrated through stacking gel, increase the voltage to 120 V and electrophorese until dye front migrates 50-75% through the gel.
Note: Native-PAGE is required to detect modification of ubiquitin. The use of cold native-PAGE running buffer, placement of gel apparatus on ice, and shortened electrophoresis distance may provide better separation and clarity of protein bands.
For detection of PR-Ubiquitin ligation and visualization of protein loading for ubiquitin modification gel, electrophorese 2.5 μl of protein ladder and 13 μl of each reaction in SDS sample buffer using an SDS-PAGE gel (4% separating gel, 12% resolving gel) and SDS-PAGE running buffer at 80 V. Once samples have migrated through stacking gel, increase the voltage to 150 V and electrophorese until dye front reaches the bottom of the gel.
Separate the gel from the casting glass and remove the stacking gel. Transfer the gel to a microwave-safe, plastic container with a lid. Pour Coomassie stain to cover gel and microwave briefly in a covered container, until boiling. Ensure not to inhale fumes when moving the container by maximizing distance from the container. Stain gel by rocking for a few hours, to overnight, at room temperature.
Note: There was some difficulty visualizing the calmodulin band using Coomassie staining. Staining temperature and time may be decreased if calmodulin can be adequately stained.
Discard stain and rinse with the destaining solution and incubate in the destaining solution for approximately 30 min to 1 h. Discard solution and repeat incubation. Repeat until protein bands are visible. If some background staining persists, allow longer water destaining in Step B5.
Rehydrate gel and destain further by incubation in ddH2O while rocking for 1-2 h. Remove water and repeat if necessary. The addition of a Kim wipe can assist in destaining and provide cleaner gel images.
After the gel is rehydrated, transfer the gel to a sheet protector and image using the gel imager. Wiping dust and staining imperfections with Kim wipe may help obtain clearer images (Figure 2).
Analyze results. The addition of phosphoribose or ADP-ribose to ubiquitin by SdeA causes a significant charge alteration relative to the overall size of the small ubiquitin protein. As a result, this alteration greatly shifts the electrophoretic mobility on a native gel, while this modification is not visualized if separating only by protein size as with SDS-PAGE. The inhibition of SdeA prevents the migration shift of ubiquitin (Figure 2A). This assay does not distinguish between the addition of ADP-ribose and further modification to PR-Ub. If assaying the PR-Ubiquitin ligation to Rab33b by SdeA, ligation is detected by incremental 8 kDa band shifts above the unmodified Rab33b band in the presence of NAD. This appearance of increased molecular weight bands corresponds to the attachment of one or multiple PR-Ub to Rab33b. Inhibition of SdeA’s PR-Ubiquitin ligase activity should decrease the intensity of the modified Rab33b compared to the uninhibited SdeA reaction (Figure 2B).

Figure 2. Reaction components required for SidJ mediated glutamylation and inhibition of SdeA. A. SidJ inhibition of the ability of SdeA to modify ubiquitin. SidJ glutamylation and SdeA modification were conducted with the reaction components and concentrations listed in Table 2, with Rab33b 1-200 replaced with reaction buffer. Reaction components were excluded as indicated. SidJ in vitro glutamylation assay was conducted for 30 min at 37 °C followed by SdeA modification for 30 min at 37 °C. Proteins were electrophoresed by native-PAGE and stained with Coomassie stain. B. SidJ inhibition of the ability of SdeA to PR-Ubiquitinate Rab33b 1-200. The reaction was conducted with concentrations listed in Table 2. SidJ in vitro glutamylation assay was conducted for 30 min at 37 °C followed by SdeA PR-Ubiquitination for 30 min at 37 °C. Proteins were electrophoresed by SDS-PAGE and the gel was stained with Coomassie stain. This figure is from the original research article (Sulpizio et al., 2019).
Recipes
Reaction Buffer
50 mM Tris pH 7.5
50 mM NaCl
Stored at room temperature
MgCl2 1 M Solution
Weigh 2.033 g of magnesium chloride, 6-hydrate and dilute to 10 ml with ddH2O
Store at room temperature
ATP 100 mM pH 7.5 Solution
Weigh 551.14 mg adenosine 5′-triphosphate disodium salt hydrate and dissolve in 8 ml of ddH2O
Adjust the pH to 7.5 with 10 M sodium hydroxide and dilute to a final volume of 10 ml with ddH2O
Aliquot and store at -80 °C
Glutamic Acid 1 M Solution
Weigh 0.936 g of L-glutamic acid monosodium salt, monohydrate and dilute to 5 ml with ddH2O
Aliquot and store at -80 °C or prepare fresh
10x SDS-PAGE Running Buffer
Store at room temperature and dilute 10-fold with ddH2O for use
Component 1 L Tris-Base 30 g Glycine 140 g SDS 10 g ddH2O To 1 L 10x Native-PAGE Running Buffer
Store at room temperature and dilute 10-fold with ddH2O for use
ddH2O To 1 LComponent 1 L Tris-Base 35 g Glycine 144 g SDS Sample Buffer
Store at room temperature, freeze aliquots -20 °C for extended storage
Component Concentration
Bromophenol Blue 0.25% (w/v)
DTT 0.5 M
Glycerol 50% (v/v)
SDS 10% (w/v)
2-Mercaptoethanol 10% (v/v)6x Native Sample Buffer
Store at room temperature
Component 50 ml
ddH2O 35 ml
Glycerol 15 ml
Bromophenol Blue 0.125 gCoomassie Stain
Store at room temperature
Component 500 ml
Methanol 225 ml
ddH2O 225 ml
Glacial Acetic Acid 50 ml
Brilliant Blue R250 1.25 gCoomassie Destaining Solution
Store at room temperature
Component 1 L
Ethanol 450 ml
ddH2O 450 ml
Glacial Acetic Acid 100 mlSDS-PAGE Gel
12% Resolving Gel
Component 8 gels Final Conc.
ddH2O 13.3 ml
30% Acrylamide/Bis Solution 16 ml 12%
1.5 M Tris pH 8.8 10 ml 375 mM
10% SDS 400 μl 0.1%
10% APS 300 μl 0.075%
TEMED 24 μl 0.06%4% Stacking Gel
Component 8 gels Final Conc.
ddH2O 11.2 ml
30% Acrylamide/Bis Solution 2.16 ml 4.14%
1.0 M Tris pH 6.8 2 ml 128 mM
10% SDS 160 μl 0.1%
10% APS 110 μl 0.07%
TEMED 16 μl 0.1%Native-PAGE Gel
8% Resolving Gel
Component 20 ml (4 Gels) Final Conc.
ddH2O 9.34 ml
1.5 M Tris, pH 8.8 5 ml 375 mM
30% Acrylamide/Bis Solution 5.34 ml 8%
10% APS 100 μl 0.05%
TEMED 20 μl 0.1%3% Stacking Gel
Component 10 ml (4 Gels) Final Conc.
ddH2O 8.32 ml
1.0 M Tris, pH 6.8 500 μl 50 mM
30% Acrylamide/Bis Solution 1.02 ml 3%
10% APS 50 μl 0.05%
TEMED 10 μl 0.1%
Acknowledgments
This work was supported by National Institute of Health (NIH) grant 5R01GM116964 (to YM), the Cornell University Harry and Samuel Mann Outstanding Graduate Student Award (to AGS), and by the NIH under Ruth L Kirschstein National Research Service Award (6T32GM008267) from the NIGMS (to MEM).
Protocol from original research article: Sulpizio, A., M. E. Minelli, M. Wan, P. D. Burrowes, X. Wu, E. J. Sanford, J.-H. Shin, B. C. Williams, M. L. Goldberg, M. B. Smolka, and Y. Mao (2019). "Protein polyglutamylation catalyzed by the bacterial calmodulin-dependent pseudokinase SidJ." eLife 8: e51162.
Competing interests
The authors declare no competing interests.
References
- Akturk, A., Wasilko, D. J., Wu, X., Liu, Y., Zhang, Y., Qiu, J., Luo, Z. Q., Reiter, K. H., Brzovic, P. S., Klevit, R. E. and Mao, Y. (2018). Mechanism of phosphoribosyl-ubiquitination mediated by a single Legionella effector. Nature 557(7707): 729-733.
- Bhogaraju, S., Bonn, F., Mukherjee, R., Adams, M., Pfleiderer, M. M., Galej, W. P., Matkovic, V., Lopez-Mosqueda, J., Kalayil, S., Shin, D. and Dikic, I. (2019). Inhibition of bacterial ubiquitin ligases by SidJ-calmodulin catalysed glutamylation. Nature 572(7769): 382-386.
- Bhogaraju, S., Kalayil, S., Liu, Y., Bonn, F., Colby, T., Matic, I. and Dikic, I. (2016). Phosphoribosylation of Ubiquitin Promotes Serine Ubiquitination and Impairs Conventional Ubiquitination. Cell 167(6): 1636-1649e1613.
- Black, M. H., Osinski, A., Gradowski, M., Servage, K. A., Pawlowski, K., Tomchick, D. R. and Tagliabracci, V. S. (2019). Bacterial pseudokinase catalyzes protein polyglutamylation to inhibit the SidE-family ubiquitin ligases. Science 364(6442): 787-792.
- Chen, Z. J. and Sun, L. J. (2009). Nonproteolytic functions of ubiquitin in cell signaling. Mol Cell 33(3): 275-286.
- Gan, N., Zhen, X., Liu, Y., Xu, X., He, C., Qiu, J., Liu, Y., Fujimoto, G. M., Nakayasu, E. S., Zhou, B., Zhao, L., Puvar, K., Das, C., Ouyang, S. and Luo, Z. Q. (2019). Regulation of phosphoribosyl ubiquitination by a calmodulin-dependent glutamylase. Nature 572(7769): 387-391.
- Haglund, K. and Dikic, I. (2012). The role of ubiquitylation in receptor endocytosis and endosomal sorting. J Cell Sci (2): 265-275.
- Havey, J. C. and Roy, C. R. (2015). Toxicity and SidJ-Mediated Suppression of Toxicity Require Distinct Regions in the SidE Family of Legionella pneumophila Effectors. Infect Immun 83(9): 3506-3514.
- Hershko, A. and Ciechanover, A. (1998). The ubiquitin system. Annu Rev Biochem 67425-479.
- Hubber, A., Kubori, T. and Nagai, H. (2013). Modulation of the ubiquitination machinery by Legionella. Curr Top Microbiol Immunol 376227-247.
- Hubber, A. and Roy, C. R. (2010). Modulation of host cell function by Legionella pneumophila type IV effectors. Annu Rev Cell Dev Biol 26261-283.
- Hurley, J. H. and Stenmark, H. (2011). Molecular mechanisms of ubiquitin-dependent membrane traffic. Annu Rev Biophys 40119-142.
- Jeong, K. C., Sexton, J. A. and Vogel, J. P. (2015). Spatiotemporal regulation of a Legionella pneumophila T4SS substrate by the metaeffector SidJ. PLoS Pathog 11(3): e1004695.
- Kotewicz, K. M., Ramabhadran, V., Sjoblom, N., Vogel, J. P., Haenssler, E., Zhang, M., Behringer, J., Scheck, R. A. and Isberg, R. R. (2017). A Single Legionella Effector Catalyzes a Multistep Ubiquitination Pathway to Rearrange Tubular Endoplasmic Reticulum for Replication. Cell Host Microbe 21(2): 169-181.
- McDade, J. E., Shepard, C. C., Fraser, D. W., Tsai, T. R., Redus, M. A. and Dowdle, W. R. (1977). Legionnaires' disease: isolation of a bacterium and demonstration of its role in other respiratory disease. N Engl J Med 297(22): 1197-1203.
- Qiu, J. and Luo, Z. Q. (2019). Methods for Noncanonical Ubiquitination and Deubiquitination Catalyzed by Legionella pneumophila Effector Proteins. Methods Mol Biol 1921267-276.
- Qiu, J., Sheedlo, M. J., Yu, K., Tan, Y., Nakayasu, E. S., Das, C., Liu, X. and Luo, Z. Q. (2016). Ubiquitination independent of E1 and E2 enzymes by bacterial effectors. Nature 533(7601): 120-124.
- Qiu, J., Yu, K., Fei, X., Liu, Y., Nakayasu, E. S., Piehowski, P. D., Shaw, J. B., Puvar, K., Das, C., Liu, X. and Luo, Z. Q. (2017). A unique deubiquitinase that deconjugates phosphoribosyl-linked protein ubiquitination. Cell Res 27(7): 865-881.
- Scheffner, M., Nuber, U. and Huibregtse, J. M. (1995). Protein ubiquitination involving an E1-E2-E3 enzyme ubiquitin thioester cascade. Nature 373(6509): 81-83.
- Shin, D., Mukherjee, R., Liu, Y., Gonzalez, A., Bonn, F., Liu, Y., Rogov, V. V., Heinz, M., Stolz, A., Hummer, G., Dotsch, V., Luo, Z. Q., Bhogaraju, S. and Dikic, I. (2020). Regulation of Phosphoribosyl-Linked Serine Ubiquitination by Deubiquitinases DupA and DupB. Mol Cell 77(1): 164-179e166.
- Sulpizio, A., Minelli, M. E., Wan, M., Burrowes, P. D., Wu, X., Sanford, E. J., Shin, J. H., Williams, B. C., Goldberg, M. L., Smolka, M. B. and Mao, Y. (2019). Protein polyglutamylation catalyzed by the bacterial calmodulin-dependent pseudokinase SidJ. Elife 8: e51162.
- Urbanus, M. L., Quaile, A. T., Stogios, P. J., Morar, M., Rao, C., , R., Evdokimova, E., Lam, M., Oatway, C., Cuff, M. E., Osipiuk, J., Michalska, K., Nocek, B. P., Taipale, M., Savchenko, A. and Ensminger, A. W. (2016). Diverse mechanisms of metaeffector activity in an intracellular bacterial pathogen, Legionella pneumophila. Mol Syst Biol 12(12): 893.
- Wan, M., Sulpizio, A. G., Akturk, A., Beck, W. H. J., Lanz, M., Faca, V. M., Smolka, M. B., Vogel, J. P. and Mao, Y. (2019). Deubiquitination of phosphoribosyl-ubiquitin conjugates by phosphodiesterase-domain-containing Legionella effectors. Proc Natl Acad Sci U S A 116(47): 23518-23526.
Article Information
Copyright
Sulpizio et al. This article is distributed under the terms of the Creative Commons Attribution License (CC BY 4.0).
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Sulpizio, A. G., Minelli, M. E. and Mao, Y. (2020). In vitro Glutamylation Inhibition of Ubiquitin Modification and Phosphoribosyl-Ubiquitin Ligation Mediated by Legionella pneumophila Effectors. Bio-protocol 10(21): e3811. DOI: 10.21769/BioProtoc.3811.
- Sulpizio, A., Minelli, M. E., Wan, M., Burrowes, P. D., Wu, X., Sanford, E. J., Shin, J. H., Williams, B. C., Goldberg, M. L., Smolka, M. B. and Mao, Y. (2019). Protein polyglutamylation catalyzed by the bacterial calmodulin-dependent pseudokinase SidJ. Elife 8: e51162.
Category
Microbiology > Microbial biochemistry > Protein > Modification
Microbiology > Microbial biochemistry > Protein > Activity
Biochemistry > Protein > Posttranslational modification
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









