- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Generation of Giant Unilamellar Vesicles (GUVs) Using Polyacrylamide Gels
Published: Vol 10, Iss 21, Nov 5, 2020 DOI: 10.21769/BioProtoc.3807 Views: 4849
Reviewed by: David PaulPhilipp A.M. SchmidpeterAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Protocol for the Isolation and Analysis of Extracellular Vesicles From Peripheral Blood: Red Cell, Endothelial, and Platelet-Derived Extracellular Vesicles
Bhawani Yasassri Alvitigala [...] Lallindra Viranjan Gooneratne
Nov 5, 2025 1500 Views

Production of Genetically Engineered Extracellular Vesicles for Targeted Protein Delivery
Leyla A. Ovchinnikova [...] Yakov A. Lomakin
Nov 5, 2025 1671 Views

A Rapid and High-Recovery Extracellular Vesicle (EVs) Isolation Technique from Blood Samples
Aryan N. Sankpal and Narendra V. Sankpal
Mar 5, 2026 65 Views
Abstract
Giant unilamellar vesicles (GUVs) are a widely used model system for a range of applications including membrane biophysics, drug delivery, and the study of actin dynamics. While several protocols have been developed for their generation in recent years, the use of these techniques involving charged lipid types and buffers of physiological ionic strength has not been widely adopted. This protocol describes the generation of large numbers of free-floating GUVs, even for charged lipid types and buffers of higher ionic strength, using a simple approach involving soft polyacrylamide (PAA) gels. This method entails glass cover slip functionalization with (3-Aminopropyl)trimethoxysilane (APTES) and glutaraldehyde to allow for covalent bonding of PAA onto the glass surface. After polymerization of the PAA, the gels are dried in vacuo. Subsequently, a lipid of choice is evenly dispersed on the dried gel surface, and buffers of varying ionic strength can be used to rehydrate the gels and form GUVs. This protocol is robust for the production of large numbers of free-floating GUVs composed of different lipid compositions under physiological conditions. It can conveniently be performed with commonly utilized laboratory reagents.
Keywords: Giant unilamellar vesicleBackground
While gentle hydration and electroformation are two of the most commonly used methods for giant unilamellar vesicle (GUV) formation, there are only a few studies that report their use for charged lipid types and in buffers of physiological ionic strength (Stein et al., 2017; Lefrançois et al., 2018). In 2009, Horger and colleagues described a simple gel-assisted swelling method for GUV generation that utilized a hybrid film of agarose gel and lipids. Unlike many other techniques, this method is capable of producing GUVs in buffers of physiological ionic strength and with charged lipid types (Horger et al., 2009). However, it was later determined that the agarose was incorporated into the GUVs, thereby affecting their mechanical properties (Lira et al., 2014). While other methods using poly(vinyl alcohol) (Weinberger et al., 2013) or dextran(ethylene glycol) (López Mora et al., 2014) have successfully developed GUVs without the incorporation of the gel into the vesicle membrane, they have not been widely utilized; this is likely due to the relatively uncommon reagents required. In our recent report (Parigoris et al., 2020), we describe another facile gel-assisted swelling method using soft polyacrylamide (PAA) gels. This technique can generate GUVs composed of both neutral and charged lipids, and under conditions of physiological ionic strength. Like the agarose gel method proposed by Horger et al. (2009), our method utilizes common chemicals present in most laboratories, and to our knowledge, does not encapsulate PAA monomers inside of the vesicles.
Materials and Reagents
5, 10, and 25 ml serological pipettes
30 mm diameter glass cover slips, round, Menzel Glaser #1 (VWR, catalog number: 631-1347 )
150 mm diameter circular Petri dishes (Fisher Scientific, catalog number: FB0875714 )
15 ml conical tubes (Corning, catalog number: 352095 )
50 ml conical tubes (Corning, catalog number: 352070 )
6-well plates (Corning, catalog number: 3516 )
Pipette tips (Appropriate for P1, P10, P20, P100, and P1000 micropipettes)
Parafilm (Sigma, catalog number: P7793 )
Large glass plate (Approximately 200 mm x 200 mm square, or larger)
Glass Drigalski spatula (can be created by melting a glass Pasteur pipette)
Razor blades
Aluminum foil
100% ethanol (Sigma, catalog number: 51976 )
Milli-Q water
Phosphate buffered saline (PBS), pH 7.4 (Thermo Fisher Scientific, catalog number: 10010001 )
Chloroform (Sigma, catalog number: C2432 )
Methanol (EMD Millipore, catalog number: 1060092500 )
(3-Aminopropyl)trimethoxysilane (Sigma, catalog number: A3648 )
Glutaraldehyde (Sigma, catalog number: G6257 )
40% acrylamide solution (Sigma, catalog number: A4058 )
2% N,N′-Methylenebisacrylamide solution (Sigma, catalog number: M1533 )
N,N,N,N-tetramethylethylenediamine (TEMED) (Bio-Rad, catalog number: 1610800 )
Ammonium persulfate (APS) (Sigma, catalog number: A3678 )
1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) lipids (Avanti, catalog number: 850375 )
1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) (Rhod-PE) lipids (Avanti, catalog number: 810158 )
Equipment
Tweezers (Fisher Scientific, catalog number: 12-000-122 )
Set of micropipettes (P1, P10, P20, P100, and P1000)
Pipet-Aid
Orbital shaker
Oven/incubator (capable of maintaining 45 °C)
Desiccator/vacuum
Vortex
Microscope with phase contrast and/or epifluorescence capabilities (Nikon, model: TS100 )
Plasma oxidizer (Diener Elektronic, FEMTO)
4 °C fridge
-20 °C freezer
Procedure
Glass cover slip functionalization
This portion of the protocol was adapted by methods described by the Engler (Tse and Engler, 2010) and Discher ( Chaudhuri et al., 2010 ) laboratories.
Place the 30 mm circular glass cover slips in a 150 mm diameter circular Petri dish. Fill the dish with 70% ethanol (enough to cover the glass cover slips), and place on an orbital shaker for a few minutes. Remove the ethanol and refill it. Repeat this step 3 times.
Note: In our hands, the best GUV formation was observed for 30 mm diameter glass cover slips. The same protocol can be adapted for smaller cover slips, and volumes can be adjusted accordingly based on the cover slip area.
Repeat Step A1 with Milli-Q water.
Remove the water from the container. Transfer the cover slips onto a holder (e.g., an old pipette tip rack) as shown in Figure 1, and place them into an oven or incubator set to 45 °C. Incubate them for 20-30 min, or until completely dry.
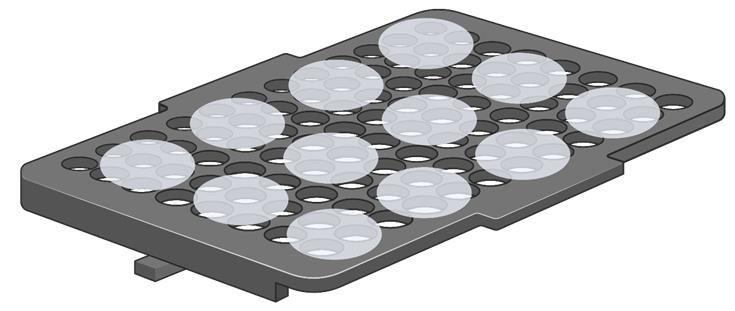
Figure 1. Placement of glass cover slips on a holder (e.g., an old pipette tip rack) in Steps A3, A13, and B11
Plasma treat the glass cover slips for 5-7 min. The upper surface will be “activated”.
While the cover slips are in the plasma cleaner, prepare 50 ml of 10% (v/v) (3-Aminopropyl)triethoxysilane (APTES) in water-free ethanol. Prepare this solution fresh each time in a conical tube. Transfer the solution to a large Petri dish.
After plasma treatment, immediately immerse the cover slips in the APTES solution for 15-30 min at room temperature using tweezers. Ensure that the cover slips are not overlapping.
Remove the APTES solution. Add ethanol to the Petri dish, and gently shake the dish to wash them. Remove the ethanol and repeat this step two more times.
Rinse each cover slip individually with ethanol on both sides. Then rinse with water, and finally PBS. Place the cover slips in a fresh Petri dish.
Note: It is very important that all of the APTES is removed. If the APTES mixes with glutaraldehyde, it produces a red/brown precipitate and the GUVs will not properly form.
Prepare 50 ml of a 2.5% (v/v) glutaraldehyde solution in PBS. Prepare this solution fresh each time.
Pour the glutaraldehyde solution into the Petri dish containing the cover slips. The “activated” side should be facing up. Incubate the cover slips at room temperature for 15-30 min and ensure that the cover slips are not overlapping.
Remove the glutaraldehyde. Add PBS to the Petri dish and gently rock the dish to rinse the cover slips. Remove the PBS and repeat this washing step two more times.
Rinse the cover slips in the Petri dish three times with Milli-Q water.
Transfer the cover slips onto a holder (e.g., an old pipette tip rack) as shown in Figure 1, and place them into an incubator set to 45 °C for 20-30 min, or until completely dry.
If not using the cover slips right away, transfer the cover slips into a Petri dish and seal it with parafilm. For optimal storage conditions, orient the cover slips with the functionalized surface facing upwards and avoid stacking the cover slips in the petri dish. Store the cover slips in a 4 °C refrigerator; they should remain functionalized for approximately 1 month.
Polyacrylamide gel preparation
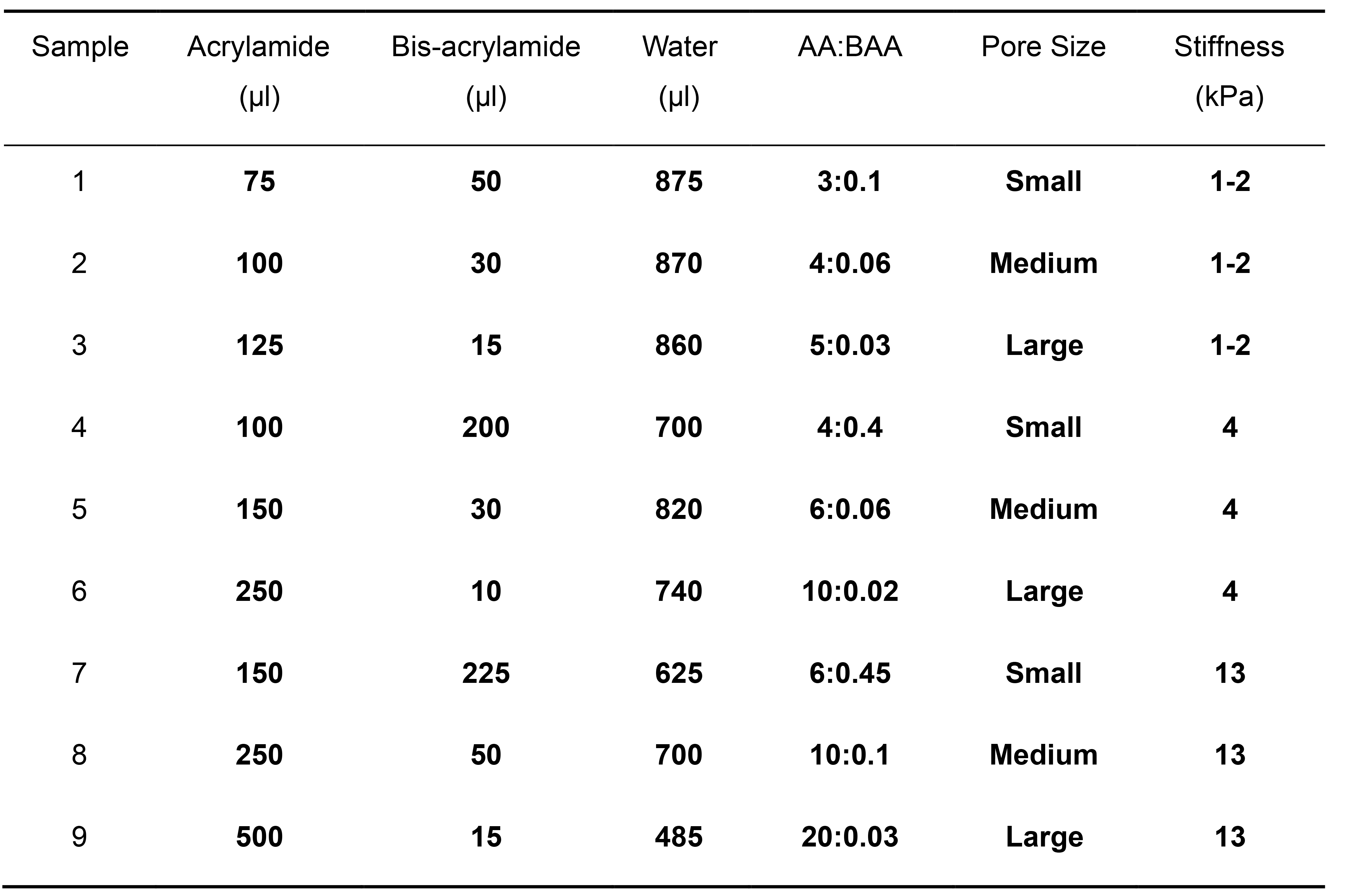
Mix the appropriate volume of acrylamide (AA), bis-acrylamide (BAA), and Milli-Q water in a 15 ml conical tube. The total volume should add to 1 ml.
Notes:
Table 1 shows nine mixes that we have confirmed to successfully produce large quantitates of GUVs. Polyacrylamide formulations were adapted from those used in Wen et al., 2014. To start, we recommend using formulation 1 or 5.
We have found that the variations in PAA stiffness and pore size do not significantly affect the concentration and size distribution of the free-floating GUVs that are generated (Parigoris et al., 2020).
Table 1. Polyacrylamide compositions used to successfully generate large populations of GUVs
Loosen the lid on the conical tube and place it in the desiccator/vacuum. Degas for approximately 30 min.
While degassing the polyacrylamide, clean a large glass plate with ethanol (approximately 200 mm x 200 mm square), followed by water, and then dry it completely.
After degassing the acrylamide and bis-acrylamide mixture, add 1 μl of Tetramethylethylenediamine (TEMED) to the polyacrylamide mixture.
Add 10 μl of 10% (w/v) ammonium persulfate (APS) to the mixture.
To make the stock solution, add Milli-Q water to 1 g of APS until a final volume of 10 ml is achieved. Then, vortex well and store aliquots at -20 °C.
Mix the solution quickly with the pipette, and briefly vortex the solution.
Add 45 μl of the polyacrylamide onto the glass plate, one droplet for each desired sample. Leave adequate spacing between each droplet.
Note: The size of the droplets depends on the diameter of the cover slip. If using a smaller cover slip, the volume of the droplet can be scaled down with the area of the cover slips.
After pipetting all of the polyacrylamide, drop one glass cover slip onto each droplet of polyacrylamide. Touch one side down, and carefully let it fall. Ensure that the functionalized surface of the cover slip is facing the droplet.
Notes:
Perform Steps B4-B8 as quickly as possible so that the polyacrylamide does not polymerize too early.
If there is an air gap between the PAA and the glass, gently move the glass cover slip back and forth to remove the bubble.
Wait 10-15 min for the polymerization of the gel.
Use the leftover polyacrylamide in the conical tube to roughly determine when it has polymerized. Pockets of air that form around the edge of the cover slips signal that the gels have sufficiently polymerized.
Wet the cover slip surface with Milli-Q water. Use a razor blade to detach the cover slips from the glass plate; be careful not to crack the cover slips during their removal.
Transfer the cover slips onto a holder (e.g., an old pipette tip rack) as shown in Figure 1, and place them into the incubator at 45 °C for approximately 20-30 min, or until completely dry. The PAA surface should be facing upwards.
GUV formation
Transfer the glass cover slips (30 mm diameter) to a 6-well plate, with one cover slip in each well. The PAA surface should be facing upwards.
Note: Adjust the well plate accordingly if you use a different size glass cover slip.
Remove the DOPC lipid solution from the -20 °C freezer and keep it on ice to prevent evaporation of the solvent.
Notes:
The lipid power should be reconstituted in a chloroform/methanol (10:1) solution to a concentration of 1.5 mM. Store all lipid solutions at -20 °C.
We recommend starting with DOPC lipids, but we have confirmed this protocol to work with a variety of neutral and charged lipid types.
To fluorescently visualize the GUVs, add 0.5% Rhod-PE lipids to the DOPC lipid solution.
Mix the lipid solution and add 20 μl to each polyacrylamide gel.
Note: The volume of lipid suspension depends on the diameter of the cover slip. If using a smaller cover slip, the volume of the droplet can be scaled down with the area of the cover slip.
Use a Drigalski spatula to evenly distribute the lipid solution across each PAA gel.
Note: Avoid touching the plastic in the plates, as the chloroform/methanol mixture will damage the plastic.
Transfer the plate to a desiccator/vacuum and degas for approximately 15 min. Leave the lid slightly open. If incorporating fluorescently labeled lipids, cover the plate with aluminum foil.
Add 750 μl of the buffer of choice to each well in order to rehydrate the lipid film.
Notes:
We have confirmed that buffers ranging from 0 mOsm to 1,000 mOsm will yield successful GUV formation. To start, use PBS or 100 mOsm sucrose as the rehydrating solution.
Add the buffer to the side of the well, and not directly over the glass/PAA.
The volume of the rehydrating solution depends on the diameter of the cover slip. If using a smaller cover slip, the volume of the droplet can be scaled down with the area of the cover slip.
Wrap the edge of the plate in parafilm and incubate at 4 °C overnight. If using fluorescently labeled lipids, cover the plate with aluminum foil. Vesicles will be formed by the morning. The GUVs will be free-floating, and can be simply transferred from the wells using a micropipette. Figure 2 shows a representative image of how the GUVs should appear with phase contrast microscopy (Figure 2A) and fluorescent microscopy (Figure 2B) if the Rhod-PE lipids are incorporated.
Notes:
We have not explored GUV formation at different temperatures; we therefore recommend overnight 4 °C incubation, though GUV formation can be seen after only a few hours.
This technique generates large numbers of free-floating GUVs (> 105 GUVs/ml). The distribution of GUV diameters ranges from ~1 μm to ~40 μm. In our measurements, the mean diameter for various PAA stiffness and porosity conditions was on the order of 10 μm (Parigoris et al., 2020).
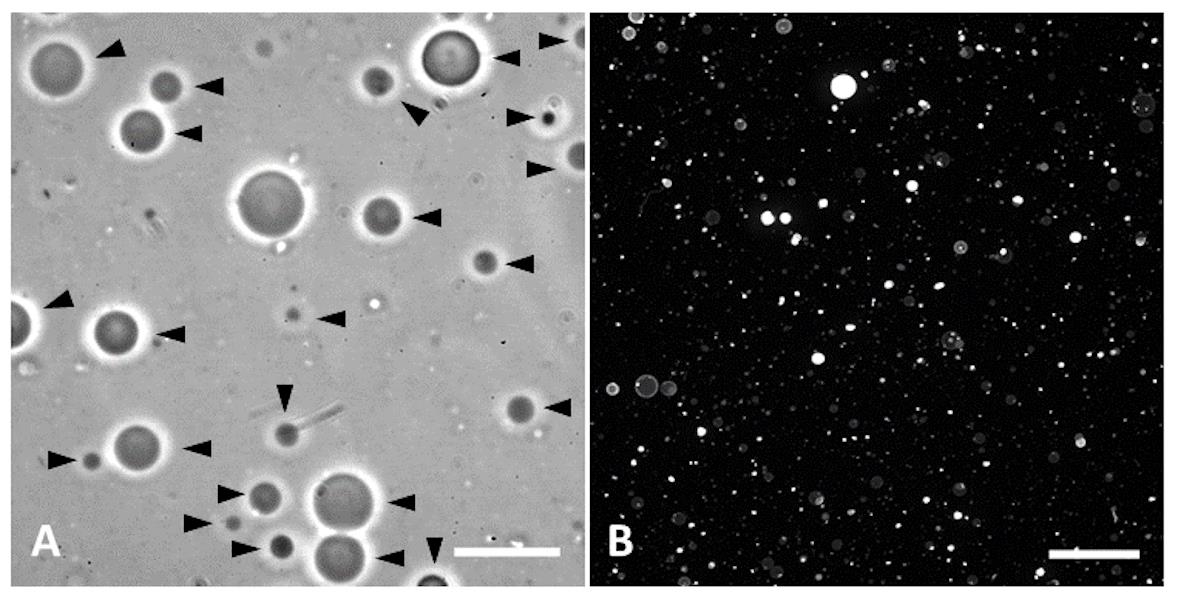
Figure 2. Representative images of GUV generation. A. Phase contrast image of DOPC GUVs. PAA-lipid gels were rehydrated in 100 mOsm sucrose, and the generated GUVs were resuspended in equiosmolar PBS (diluted to 100 mOsm) to aid in their visualization. Scale bar represent 20 μm. Black arrows point to individual GUVs. B. Maximum Z-projection of fluorescent confocal stack of DOPC GUVs formed in PBS. 0.5% Rhod-PE were added to the lipid mixture to aid in their visualization. Scale bar represents 100 μm.Acknowledgments
This work was supported by the Whitaker International Fellowship to EP. The authors would like to thank Professor Jess G. Snedeker for supporting this project. Figure 1 was generated with Biorender.
Competing interests
The authors declare no competing interests.
References
- Chaudhuri, T., Rehfeldt, F., Sweeney, H. L. and Discher, D. E. (2010). Preparation of collagen-coated gels that maximize in vitro myogenesis of stem cells by matching the lateral elasticity of in vivo muscle. Methods Mol Biol 621185-202.
- Horger, K. S., Estes, D. J., Capone, R. and Mayer, M. (2009). Films of agarose enable rapid formation of giant liposomes in solutions of physiologic ionic strength. J Am Chem Soc 131(5): 1810-1819.
- Lefrançois, P., Goudeau, B. and Arbault, S. (2018). Electroformation of phospholipid giant unilamellar vesicles in physiological phosphate buffer. Integr Biol (Camb) 10(7): 429-434.
- Lira, R. B., Dimova, R. and Riske, K. A. (2014). Giant unilamellar vesicles formed by hybrid films of agarose and lipids display altered mechanical properties. Biophys J 107(7): 1609-1619.
- López Mora, N., Hansen, J. S., Gao, Y., Ronald, A. A., Kieltyka, R., Malmstadt, N. and Kros, A. (2014). Preparation of size tunable giant vesicles from cross-linked dextran(ethylene glycol) hydrogels. Chem Commun (Camb) 50(16): 1953-1955.
- Parigoris, E., Dunkelmann, D. L., Murphy, A., Wili, N., Kaech, A., Dumrese, C., Jimenez-Rojo, N. and Silvan, U. (2020). Facile generation of giant unilamellar vesicles using polyacrylamide gels. Sci Rep 10(1): 4824.
- Stein, H., Spindler, S., Bonakdar, N., Wang, C. and Sandoghdar, V. (2017). Production of isolated giant unilamellar vesicles under high salt concentrations. Front Physiol 863.
- Tse, J. R. and Engler, A. J. (2010). Preparation of hydrogel substrates with tunable mechanical properties. Curr Protoc Cell Biol Chapter 10: Unit 10.16.
- Weinberger, A., Tsai, F. C., Koenderink, G. H., Schmidt, T. F., Itri, R., Meier, W., Schmatko, T., Schroder, A. and Marques, C. (2013). Gel-assisted formation of giant unilamellar vesicles. Biophys J 105(1): 154-164.
- Wen, J. H., Vincent, L. G., Fuhrmann, A., Choi, Y. S., Hribar, K. C., Taylor-Weiner, H., Chen, S. and Engler, A. J. (2014). Interplay of matrix stiffness and protein tethering in stem cell differentiation. Nature Mater 13(10): 979-987.
Article Information
Copyright
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Parigoris, E., Dunkelmann, D. L. and Silvan, U. (2020). Generation of Giant Unilamellar Vesicles (GUVs) Using Polyacrylamide Gels. Bio-protocol 10(21): e3807. DOI: 10.21769/BioProtoc.3807.
Category
Biochemistry > Lipid > Membrane lipid
Cell Biology > Organelle isolation > Extracellular vesicle
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











