- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Murine Acute Pneumonia Model of Pseudomonas aeruginosa Lung Infection
Published: Vol 10, Iss 21, Nov 5, 2020 DOI: 10.21769/BioProtoc.3805 Views: 4645
Reviewed by: Kristin L. ShinglerElena Jordana-LluchAyush Ranawade

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

HS–GC–MS Method for the Diagnosis of IBD Dynamics in a Model of DSS-Induced Colitis
Olga Yu. Shagaleeva [...] Natalya B. Zakharzhevskaya
Mar 20, 2025 3108 Views

In Vitro Co-culture of Bacterial and Mammalian Cells to Investigate Effects of Potential Probiotics on Intestinal Barrier Function
Ajitpal Purba [...] Dulantha Ulluwishewa
Jun 20, 2025 2515 Views

In Silico Prediction and In Vitro Validation of Bacterial Interactions in the Plant Rhizosphere Using a Synthetic Bacterial Community
Arijit Mukherjee [...] Sanjay Swarup
Nov 5, 2025 1655 Views
Abstract
Animal infection models play significant roles in studying bacterial pathogenic mechanisms, host pathogen interaction as well as evaluating drug and vaccine efficacies. We have been utilizing an acute pneumonia model to study bacterial colonization in lungs and assess virulence to the host by determination of bacterial loads and survival assays, as well as examine the bacterial gene expression in vivo. Additionally, the host's immune response to the pathogen can be explored through this infection model.
Background
Acute pneumonia is defined as an acute infection of the lungs by microbial pathogens. Hospital-acquired pneumonia, in particular, is often caused by multi-drug resistant pathogens and is difficult to treat. Common bacteria that cause pneumonia are Pseudomonas aeruginosa, Streptococcus pneumoniae, group A Streptococcus, Acinetobacter baumannii, Klebsiella pneumoniae,Staphylococcus aureus and Mycoplasma pneumonia (Ravi Kumar et al., 2018 ). To develop effective prevention and treatment strategies, it is critical to understand how bacteria sense and adapt to host in vivo environment and counteract host immune clearance. Currently, there are three types of pneumonia models: One hit acute pneumonia model, Ventilator-associated pneumonia model, and Agar-bead pneumonia model (Bielen et al., 2017). The model mentioned in this article belongs to the one hit acute pneumonia model. There are two methods for the infection, one is to inject bacterial suspension directly into the trachea or lungs. Here, we reported the other method in which bacteria are inoculated through nostrils, which is relatively easy to perform.
P. aeruginosa is a Gram negative opportunistic pathogenic bacterium that causes a variety of acute and chronic infections in human. It is one of the major pathogens that cause lung infections in patients with cystic fibrosis, chronic obstructive pulmonary disease and compromised immunity (Talwalkar and Murray, 2016). To study the virulence factors and gene expression of P. aeruginosa in host, we utilized gene expression (Wu et al., 2012). Additionally, this model is suitable for examining the immune response of host (Tian et al., 2019). We have also applied this model to study the infections by K. pneumoniae and A. baumannii.
Materials and Reagents
Plastic EP tubes (1.5 ml, 2 ml, 4 ml, G SERVICEBIO, catalog numbers: EP-150-M , EP-200-M , EP-400-M )
Syringe (1 ml, Anhui JINHUAN Co., Ltd, catalog number: 14001 )
Pipette tips (20, 200, and 1,000 μl clear tips, Tianjin HUAXIN Co., Ltd, catalog number: HY130 )
Six-week-old female BALB/c mice (Beijing Vital River Laboratory Animal Technology Co., Ltd)
Bacterial culture (P. aeruginosa wild type strain PA14 [ Liberati et al., 2006 ])
75% ethanol (Tianjin Chemicals Co., Ltd, catalog number: A1022 ). Diluent: double distilled water.
7.5% chloral hydrate (Tianjin Chemicals Co., Ltd, catalog number: 2223 ). Diluent: double distilled water
0.05 M Ethylenediaminetetraacetic acid (EDTA) disodium salt solution, pH 8.0 (Beijing Dingguo Biotech Co., Ltd, catalog number: LE568-1KG ). Diluent: double distilled water
Carbon dioxide (Tianjin Baisida Co., Ltd, catalog number: N14 )
1% sterilized peptone (Oxiod Co., Ltd, catalog number: LP0037B ). Diluent: double distilled water.
Trizol (BioFroxx Co., Ltd, catalog number: 15596026 )
Isopropanol (Tianjin Chemicals Co., Ltd, catalog number: A1079 )
Chloroform (Tianjin Chemicals Co., Ltd, catalog number: A1008 )
Phosphate-buffered saline (PBS), pH 7.2 (see Recipes)
NaCl (Invitrogen, catalog number: 24740011 )
KCl (Aladdin, catalog number: P112134-12×500g)
Na2HPO4·7H2O (Tianjin Chemicals catalog number: W2007)
KH2PO4 (Macklin, catalog number: M822553-100g)
LB broth (see Recipes)
NaCl (Invitrogen, catalog number: 24740011 )
Tryptone (Abbexa, catalog number: abx082523-500g )
Yeast extract (Abbexa, catalog number: abx082370-500g )
Equipment
Pipette (GILSON, catalog number: F167350 )
Incubator (Shanghai Yihen Co., Ltd, catalog number: 2005A )
-80 °C freezer
Centrifuge (Eppendorf, model: 5810 )
Vortex (BioExpress, GeneMate, catalog number: S-3200-1 )
Spectrophotometer (BIORAD, model: Smart Spec plus )
Surgical scissors
Fine Forceps
I.V. Catheter (BD, model: Angiocath 1.3 x 48 mm )
Electronic homogenizer (Shanghai JIANLI Co., Ltd, catalog number: e76 )
Software
GraphPad
Procedure
Time line of the procedures (Figure 1):
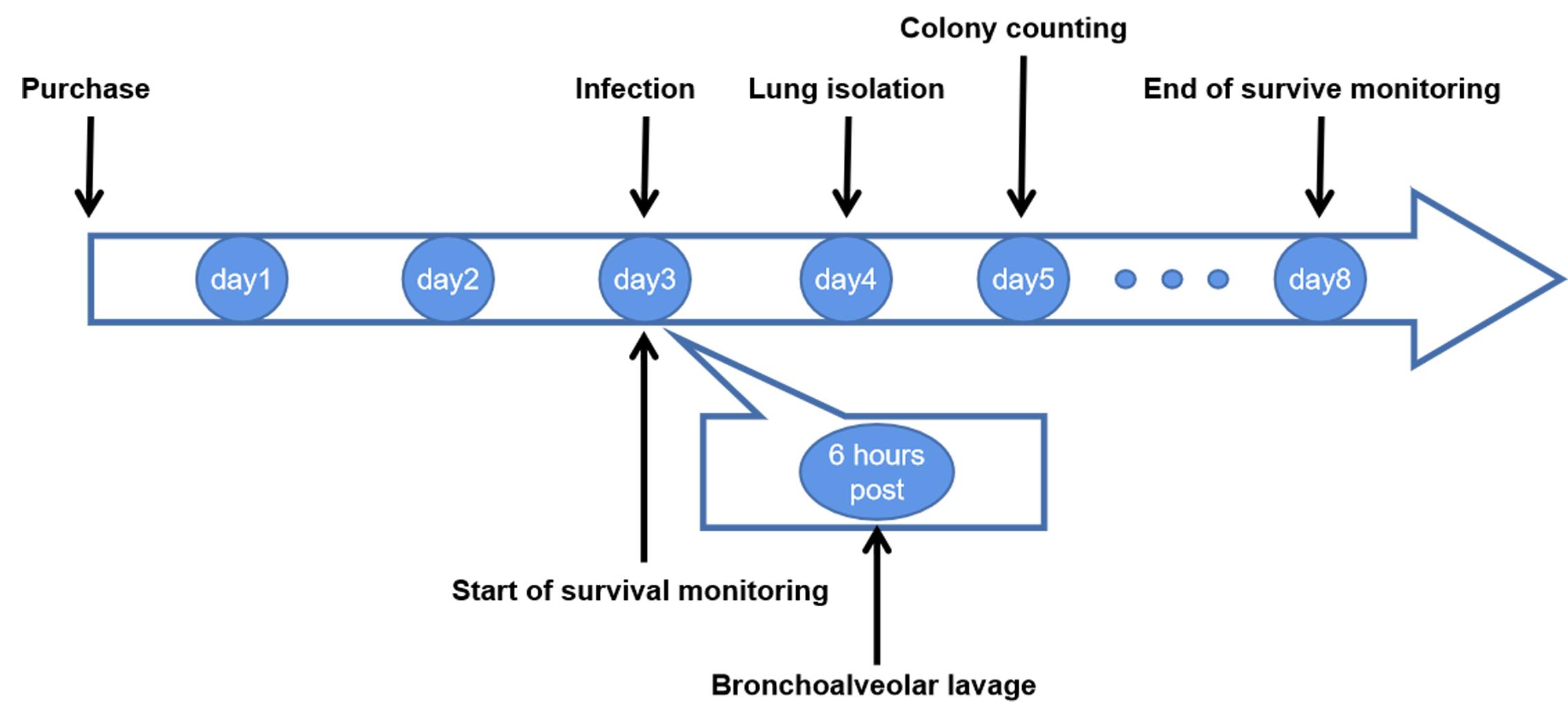
Figure 1. Time line of the procedures
Infection
Specific pathogen free (SPF) six-week-old female BALB/c mice are purchased from Beijing Vital River. After arrival, the mice are kept in animal facility for three to five days for acclimatization ( Wu et al., 2012 ).
Streak the P. aeruginosa reference stain PA14 stored at -80 °C on a LB plate. Incubate the plate at 37 °C overnight.
Inoculate a single colony on the plate into LB liquid medium and culture the bacteria at 37 °C for 16 h with shaking at 200 rpm.
Dilute the bacterial culture 1:100 into 3 ml fresh LB broth.
Culture the bacteria at 37 °C with shaking at 200 rpm for about 3 h until the OD600 reached 1.0.
Collect 1 ml of the bacteria by centrifugation at 10,000 x g for 1 min at room temperature in a 1.5 ml EP tube. Discard the supernatant and resuspend the bacteria with 1 ml PBS at room temperature. Centrifuge the bacteria at 10,000 x g for 1 min at room temperature. Resuspend the bacteria with 1 ml PBS.
Dilute the bacteria to the desired concentration in PBS (e.g., 1 x 109 CFU per ml of PA14).
Anesthetize the mouse with an intraperitoneal injection of 7.5% chloral hydrate (100 µl per 20 g mouse).
Flip the mouse onto its back and place it in the palm of your hand.
Press the toe lightly to make sure the mouse has been anesthetized. Anesthetized mouse does not respond to the pressing.
Hold the mouse’s body at a 45-degree angle.
Place thumb lightly on the mouse's jaw to force the mouse to breathe through nose (Figure 2A).
Take 10 μl of the bacteria suspension or PBS as control with a 20 μl pipette. Put the tip close to one of the nostrils of the mouse. Slowly pipette a drop of bacteria suspension onto the nostril. After the liquid is inhaled into the nostril, pipette the next drop until all the 10 μl is delivered. Repeat the process with another 10 μl of the bacteria suspension on the other nostril (Figure 2B, Video 1). During the process, pauses might be needed for restoring the normal breathing of the mouse.
Lay the mouse on its back in the cage.
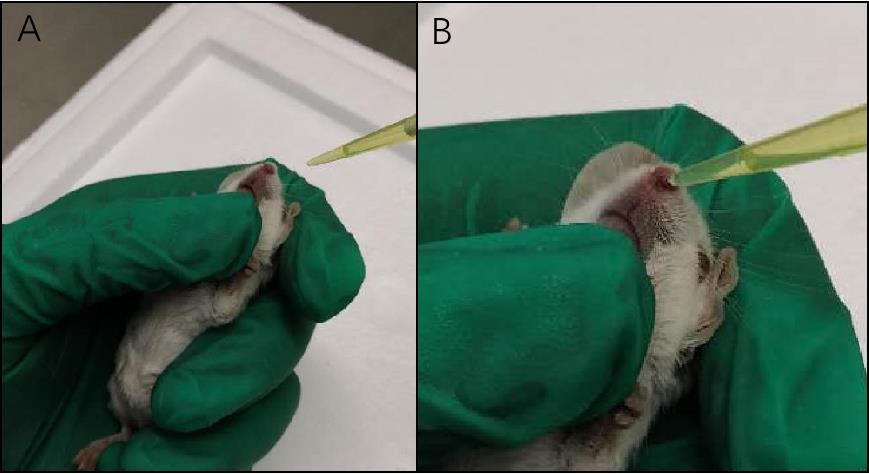
Figure 2. Key steps of inoculation. A. Mouse on its back with jaw being pressed lightly. B. Pipette bacteria suspension onto the nostril.Video 1. An illustration of the process of the murine acute pneumonia model of P. aeruginosaDetermination of bacterial load
At desired time point post infection (e.g., 12 h), fill the closed container with carbon dioxide to sacrifice the mice by asphyxia.
Spread the forelimbs of the mouse and affix to a working surface with syringe needles.
Using fine scissors, cut the skin from the intersection of the abdomen and the chest to the leading edge of chest to open the chest cavity. Then cut the ribs on each side to expose the lung.
Isolate the lung with a scissor and immerse the lung in 1 ml 1% sterilized peptone in a 4 ml EP tube (Steps B2-B4 are shown in Video 2).
Video 2. Lung isolating for determination of the colonization of P. aeruginosa in the murine acute pneumonia modelGrind the lung with an electronic homogenizer until it is completely homogenized (no particles larger than 1 mm in diameter). For our homogenizer and mice, the homogenization parameters of 65 HZ homogenize 20 s for 10 times with 10-second intervals work. If the readers uses a different instrument type or mouse type/weight, we recommend that new parameters should be tested to achieve completed homogenization with the shortest homogenization time.
Centrifuge the homogenate at 10,000 x g for 1 min at room temperature and transfer the supernatant to a new EP tube.
Add 50 μl of the supernatant to 200 μl LB in a 1.5 ml EP tube and mix well by vortex for 10 s. Transfer 50 μl of the acquired diluent to another 200 μl LB, repeat the dilution four to five times.
Spot 15 μl of the last 3 diluent onto square LB plates and immediately tilt the plate to a 45 degree angle until the fluid flows about 2-3 cm long. Incubate the plates at 37 °C for 12 h before colony counting.
After incubate for 12 h, plates should look like the Figure 3 shown. All colonies in each column are counted to calculate the bacteria load in an individual lung. For example, colonies in red box are counted as 85. If the sample is diluted 4 times, the colonization in this lung is 46 x 54 x (1,000/15) =1916667 CFU/lung.
Analyze the results with the GraphPad software (Figure 4A). Every group should contain at least 6 mice.
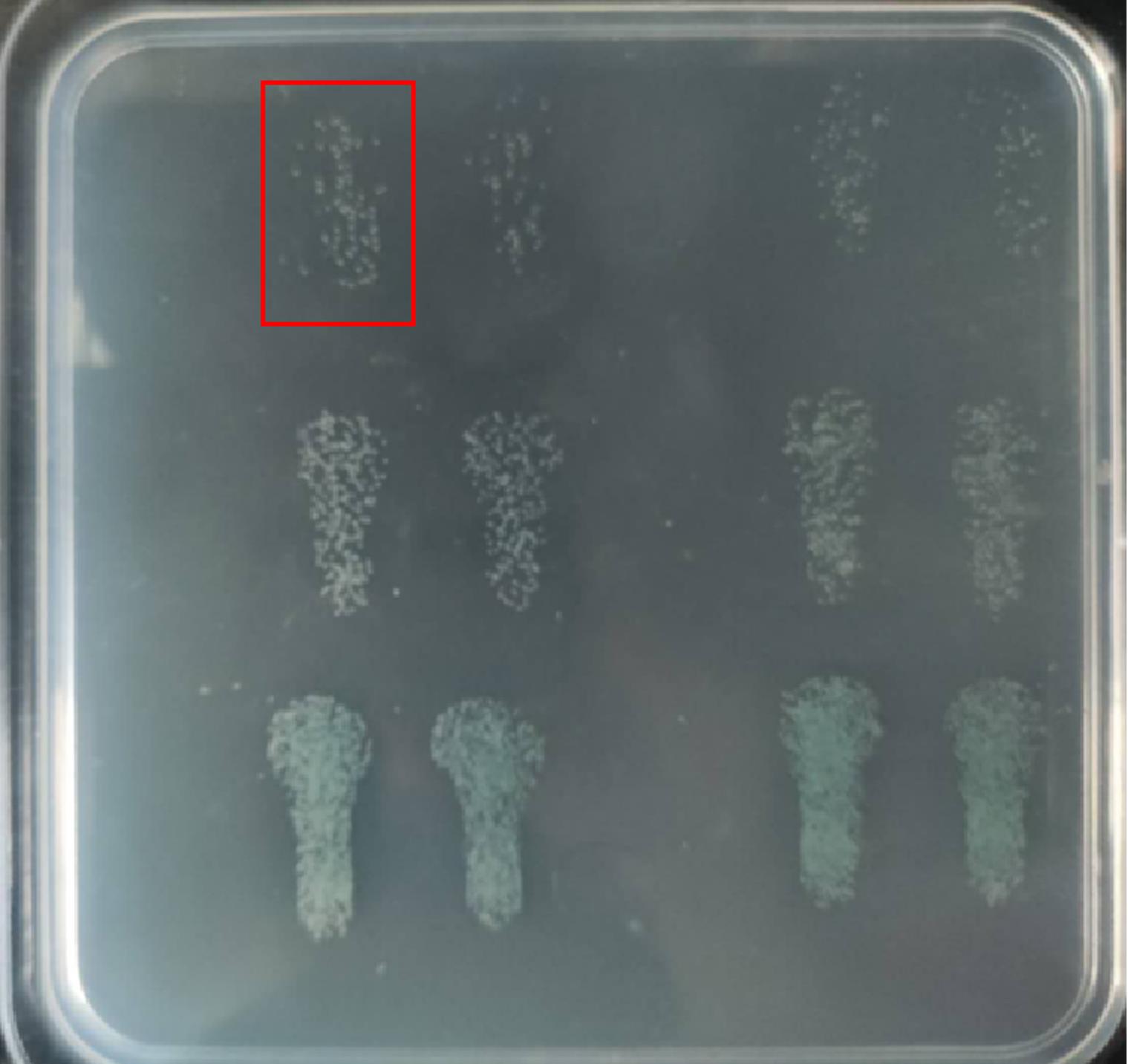
Figure 3. Illustration of plate for countingSurvival assays
After inoculation of the bacteria, check the mice at least twice a day, preferably in the morning and evening, record the survival number.
Monitor the mice for 5 days post infection, make a survival curve by the GraphPad software (Figure 4B). Every group should contain at least 8 mice.
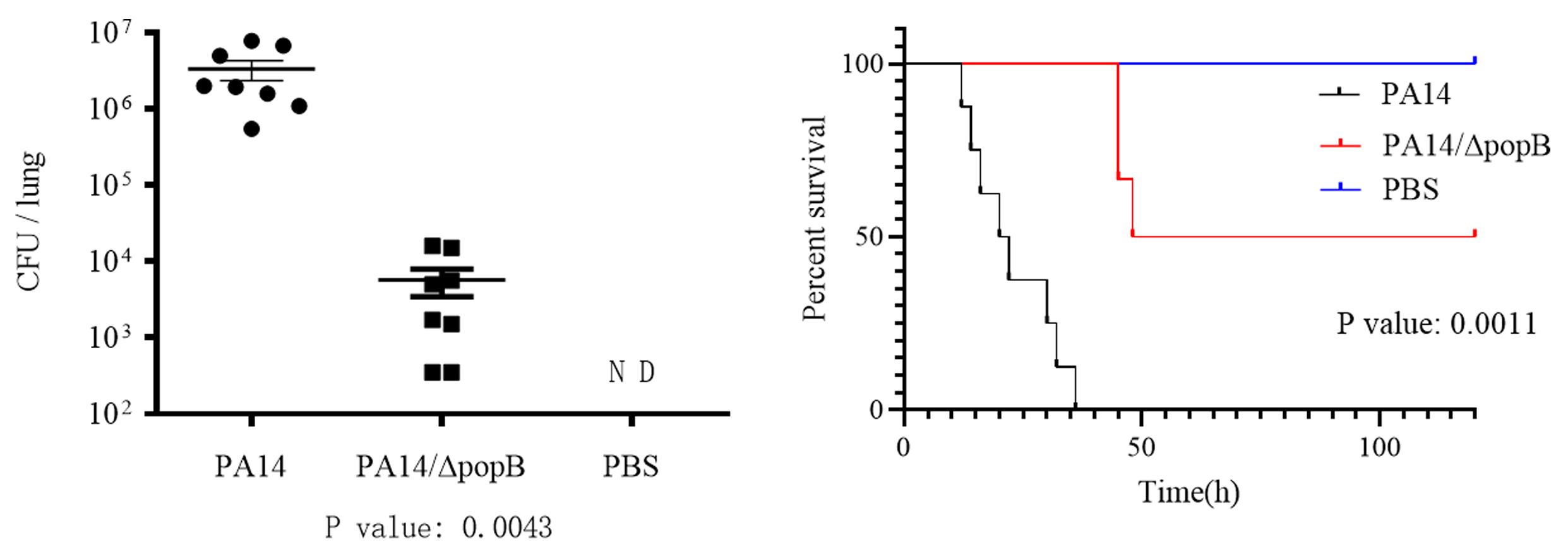
Figure 4. Data presentation of (A) Bacterial load determination and (B) Survival assays. GraphPad is used for data analyses. Bars represent medians, and error bars represent standard errors (SEM). P value < 0.05 compared to wild type PA14 by the Kruskal-Wallis with Dunn’s multiple comparison test. PA14: wild type P. aeruginosa, PA14ΔpopB: PA14 with the popB gene deleted, PBS: phosphate buffer saline buffer as a negative control, ND: not detected.RNA isolation from bacteria in bronchoalveolar lavage fluid (BALF)
Collection of the BALF
At desired time point (6 h for PA14), sacrifice the mouse with CO2. Fix the mouse on a working surface as described in Procedure B.
Using fine scissors, cut from the chest to the base of the intersection of the chin and neck. Open the chest to expose the lung and the trachea.
Carefully clear the tissue around the trachea by using a fine forceps until the trachea is exposed, be careful not to cut any blood vessel.
With the mouse head toward you, carefully insert an I.V. catheter into trachea from the top down (Figure 5A, Video 3).
Once the I.V. Catheter is half an inch into the trachea, carefully pull out the metal part of the I.V. Catheter.
Connect the syringe with 1 ml of 0.05 mM EDTA in PBS to the end of the I.V. Catheter.
Carefully and slowly inject the solution into the lungs. The lungs will expand as a result, then wait 1 min prior to drawing back (Figure 5B, Video 3).
Collect the liquid to the 2 ml tube.
Repeat Steps D1e-D1g once with another 1 ml of 0.05 mM EDTA in PBS solution.
Keep the BALF on ice until all the mice have been finished.
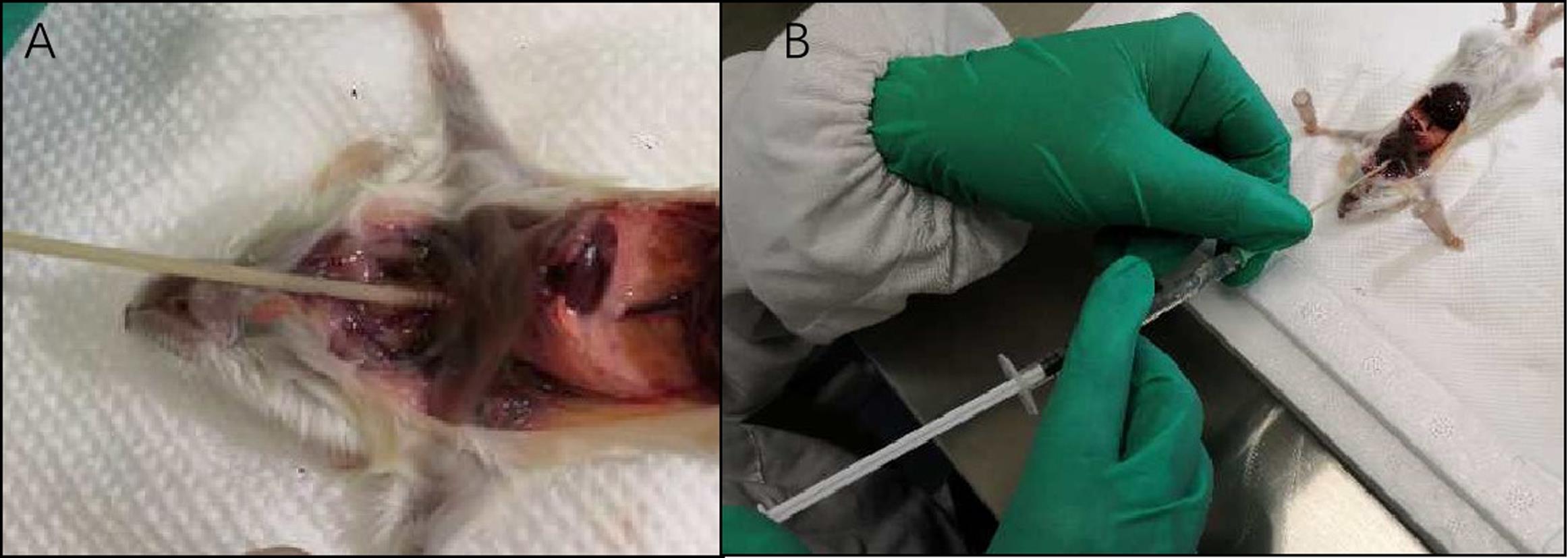
Figure 5.Key steps of collection of bronchoalveolar lavage fluid (BALF). A. Insertion of an I.V. Catheter into trachea. B. Injection of the solution into lung through the trachea.
Video 3. Lung lavage for mRNA isolation in the the murine acute pneumonia model
RNA extraction
Take 20 μl of the BALF for bacteria counting by serial dilution and plating (refer to Procedure B).
Collect the remaining bacteria in the BALF by centrifugation at 10,000 x g for 1 min at 4 °C in a 1.5 ml EP tube.
Immediately resuspend the bacteria with 1 ml Trizol for RNA isolation.
Incubate the homogenized sample for 5 min at room temperature to permit complete dissociation of the nucleoprotein complex.
Add 0.2 ml of chloroform per 1 ml of TRIzol Reagent used for homogenization. Cap the tube securely.
Shake tube vigorously by hand for 15 s.
Incubate for 2-3 min at room temperature.
Centrifuge the sample at 12,000 x g for 15 min at 4 °C.
Note: The solution are separated into a lower red phenol-chloroform phase (protein), an interphase (DNA), and a colorless upper aqueous phase (RNA). RNA remains exclusively in the aqueous phase. The upper aqueous phase is ~50% of the total volume.
Take the aqueous phase of the sample avoid drawing any of the interphase or organic phase into the pipette.
Place the aqueous phase into a new tube.
Add 0.5 ml of 100% isopropanol to the aqueous phase, per 1 ml of TRIzol Reagent used for homogenization. Mix vigorously.
Incubate at room temperature for 10 min.
Centrifuge at 12,000 x g for 10 min at 4 °C.
Carefully remove the supernatant.
Wash the precipitate twice with 0.5 ml 75% ethanol.
Put the tubes at room-temperature for 5-10 min uncovering the lid.
Add 25-50 μl H2O to the precipitate and incubate at room temperature to dissolve the RNA.
The purified RNA can be used for q-RT PCR or RNA sequencing.
Recipes
Phosphate-buffered saline (PBS), pH 7.2
137 mM NaCl
2.7 mM KCl
4.3 mM Na2HPO4·7H2O
1.4 mM KH2PO4
LB broth
NaCl 5 g/L
Tryptone 5 g/L
Yeast extract 10 g/L
Acknowledgments
We thank members of the laboratory for improving on this technique over the years. This work was supported by the National Science Foundation of China (31900115).
Competing interests
We declare no conflict of interest or competing interests.
All animal studies have been approved by the institutional animal care and use committee of the College of Life Sciences of Nankai University (permit number NK-04-2012).
Ethics
All animal studies have been approved by the institutional animal care and use committee of the College of Life Sciences of Nankai University (permit number NK-04-2012).
References
- Bielen, K., Jongers, B., Malhotra-Kumar, S., Jorens, P. G., Goossens, H. and Kumar-Singh, S. (2017). Animal models of hospital-acquired pneumonia: current practices and future perspectives. Ann Transl Med 5(6): 132.
- Liberati, N. T., Urbach, J. M., Miyata, S., Lee, D. G., Drenkard, E., Wu, G., Villanueva, J., Wei, T. and Ausubel, F. M. (2006). An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc Natl Acad Sci U S A 103(8): 2833-2838.
- Ravi Kumar, S., Paudel, S., Ghimire, L., Bergeron, S., Cai, S., Zemans, R. L., Downey, G. P. and Jeyaseelan, S. (2018). Emerging Roles of Inflammasomes in Acute Pneumonia. Am J Respir Crit Care Med 197(2): 160-171.
- Talwalkar, J. S. and Murray, T. S. (2016). The Approach to Pseudomonas aeruginosa in Cystic Fibrosis. Clin Chest Med 37(1): 69-81.
- Tian, Z., Cheng, S., Xia, B., Jin, Y., Bai, F., Cheng, Z., Jin, S., Liu, X. and Wu, W. (2019). Pseudomonas aeruginosa ExsA Regulates a Metalloprotease, ImpA, That Inhibits Phagocytosis of Macrophages. Infect Immun 87(12): e00695-19.
- Wu, W., Huang, J., Duan, B., Traficante, D. C., Hong, H., Risech, M., Lory, S. and Priebe, G. P. (2012). Th17-stimulating protein vaccines confer protection against Pseudomonas aeruginosa pneumonia. Am J Respir Crit Care Med 186(5): 420-427.
Article Information
Copyright
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Pan, X. and Wu, W. (2020). Murine Acute Pneumonia Model of Pseudomonas aeruginosa Lung Infection. Bio-protocol 10(21): e3805. DOI: 10.21769/BioProtoc.3805.
Category
Microbiology > in vivo model > Bacterium
Microbiology > Microbe-host interactions > Bacterium
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link