- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Colorimetric RT-LAMP Methods to Detect Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2)
Published: Vol 10, Iss 21, Nov 5, 2020 DOI: 10.21769/BioProtoc.3804 Views: 5856
Reviewed by: Alba BlesaDay-Yu ChaoThibaud T. Renault

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

A Protocol for Simple, Rapid, and Direct Detection of SARS-CoV-2 from clinical samples, using Reverse Transcribed Loop-Mediated Isothermal Amplification (RT-LAMP)
Rawi Naddaf [...] Naama Geva-Zatorsky
Oct 20, 2020 5223 Views

A Novel PCR-Based Methodology for Viral Detection Utilizing Mechanical Homogenization
Zachary P. Morehouse [...] Rodney J. Nash
Mar 5, 2022 2761 Views

Direct RNA Sequencing of Foot-and-mouth Disease Virus Genome Using a Flongle on MinION
Lizhe Xu [...] Bonto Faburay
Jun 20, 2024 2144 Views
Abstract
Standard diagnostic methods of Coronavirus Disease 2019 (COVID-19) rely on RT-qPCR technique which have limited point-of-care test (POCT) potential due to necessity of dedicated equipment and specialized personnel. LAMP, an isothermal nucleic acid amplification test (NAAT), is a promising technique that may substitute RT-qPCR for POCT of genomic materials. Here, we provide a protocol to perform reverse transcription LAMP targeting SARS-CoV-2. We adopted both real-time fluorescence detection and end-point colorimetric detection approaches. Our protocol would be useful for screening diagnosis of COVID-19 and be a baseline for development of improved POCT NAAT.
Keywords: SARS-CoV-2Background
Fast and sensitive detection of SARS-CoV-2, the etiologic agent of COVID-19, is important to control current pandemic situation as it enables early detection, isolation and treatment as well as monitoring screening. Current standard methods for detection of SARS-CoV-2 utilize RT-qPCR as World Health Organization recommends (WHO, 2020) However, proper performance of RT-qPCR diagnosis requires high profile facilities and specialists, often not available in the hospital/sampling places, thus, lacking POCT suitability.
Isothermal amplification methods are developed to accomplish diagnosis by nucleic acid detection in various point-of-care as it can be performed with relatively simple instruments (Niemz et al., 2011). Loop-mediated isothermal amplification (LAMP) is one of such isothermal NAAT (Notomi et al., 2000). Amplification by LAMP reaction can be observed through fluorescent dye with real-time PCR instruments (Oscorbin et al., 2016), or through cost effective colorimetric detection methods (Goto et al., 2009; Miyamoto et al., 2015; Tanner et al., 2015). Indeed reverse transcription LAMP (RT-LAMP) may be considered a promising tool as several other groups are employing this technique for SARS-CoV-2 detection (Yan et al., 2020; Zhang et al., 2020).
Here, we present our protocol used to develop RT-LAMP assays targeting SARS-CoV-2. Candidate primer sets are designed using web based tool PrimerExplorerV5 (https://primerexplorer.jp/e/) targeting SARS-CoV-2 specific regions compare to SARS-CoV and SARS-CoV-2 Nucleocapsid. The primer sets are screened for limit of detection and threshold time measured through real time fluorescent method. Below illustrated protocol includes RNA standard preparation, titration of cultured viral RNA and LAMP reaction which are used during primer screening (Figure 1). Optimized RT-LAMP condition for finally selected two primer sets, namely “Nsp3_1-61” and “Nsp3_2-24”, are representatively provided. We also adapted leuco crystal violet (LCV) colorimetric detection method optimized for Bst 3.0 buffer system (Miyamoto et al., 2015). A commercially available colorimetric RT-LAMP premix uses pH sensitive dye so that the premix is not compatible with viral genome extraction methods which affect pH of weakly buffered reaction solution (Lalli et al., 2020). However, LCV method is compatible with such viral genome extraction methods because the color change depends on dsDNA product.
Figure 1. Protocol overview
Materials and Reagents
LightCycler® 8-Tube Strips, Clear (Roche, catalog number: 06327672001)
LightCycler® 480 Multiwell Plate 96, Clear (Roche, catalog number: 05102413001)
AccuPower PCR PreMix (-dye) kit (Bioneer, catalog number: K-2016 )
Agarose (Bio Basic, catalog number: D0012 )
50x TAE (Biosesang, catalog number: T2002 )
MEGAscriptTM T7 Transcription Kit (Invitrogen, catalog number: AM1334 )
Formaldehyde, 37% w/w aq. Soln. (Alfa Aesar, catalog number: A16163 )
10x MOPS (Bioneer, catalog number: C-9031 )
SYBRTM Green II RNA Gel Stain (Invitrogen, catalog number: S7564 )
RiboRuler Low Range RNA Ladder (Thermo Scientific, catalog number: SM1831 )
QuantiFluor® RNA System (Promega, catalog number: E3310 )
1 M Tris-HCl, pH 7.5 (Biosesang, catalog number: T2016-7.5 )
0.5 M EDTA, pH 8 (Biosesang, catalog number: E2002 )
QIAamp Viral RNA Mini Kit (Qiagen, catalog number: 52906 )
Luna® Universal Probe One-Step Reaction Mix (NEB, catalog number: E3006 )
DEPC treated water (Biosesang, catalog number: W2004 )
WarmStart® Colorimetric LAMP 2x Master Mix (NEB, catalog number: E1800 )
SYTOTM 9 (Invitrogen, catalog number: S34854 )
Bst 3.0 DNA polymerase (NEB, catalog number: M0374 , The product includes Isothermal Amplification Buffer II and Magnesium Sulfate solution.)
dNTP Mixture, 10 mM ea. (Enzynomics, catalog number: N002 )
SuperScriptTM IV Reverse Transcriptase (Invitrogen, catalog number: 18090050 )
Crystal Violet (Sigma, catalog number: C0775 )
Sodium Sulfite (Sigma, catalog number: S0505 )
β-Cyclodextrin (Sigma, catalog number: C4767 )
pET21a plasmid containing SARS-CoV-2 Envelope gene (Bionics, custom order)
T7 promoter primer (5′-AATACGACTCACTATAG-3′) and T7 terminator primer (5′-GCTAGTTATTGCTCAGCGG-3′) (Macrogen) (Table 1)
Denaturing agarose gel (1%, 100 ml) (see Recipes)
20 μM SYTO 9 (see Recipes)
5x LCV solution (see Recipes)
Table 1. Primer sequences used for RT-qPCR and RT-LAMP

Equipment
Mupid-One (ADVANCE, catalog number: AD160 )
ChemiDocTM Touch Imaging System (Bio-Rad, catalog number: 1708370 )
QuantusTM Fluorometer (Promega, catalog number: E6150 )
LightCycler® 96 Instrument (Roche, catalog number: 0 5815916001 )
Software
Polynucleotide Molecular Weight Calculator (Developed by Andrew Staroscik, http://scienceprimer.com/nucleotide-molecular-weight-calculator)
LightCycler® 96 Software (Roche)
Procedure
Preparation of in vitro transcribed RNA standards for SARS-CoV-2 E gene
Prepare PCR product as template for in vitro transcription: Run PCR with AccuPower® PCR PreMix (-dye) kit as manufacturer’s instructions. Use pET21a plasmid containing SARS-CoV-2 Envelope gene as template and T7 promoter primer and T7 terminator primer.
Perform in vitro transcription using MEGAscriptTM T7 Transcription Kit as manufacturer’s instructions.
Confirm RNA product by subjecting a portion to formaldehyde-MOPS denaturing agarose gel electrophoresis. The same volume of RNA loading buffer II included in MEGAscriptTM T7 Transcription Kit was added and heated for 10 min at 65 °C for sampling. Post-stain gel with 2x SYBR green II in 1x TAE and obtain image using ChemiDocTM Touch Imaging System (Figure 2).
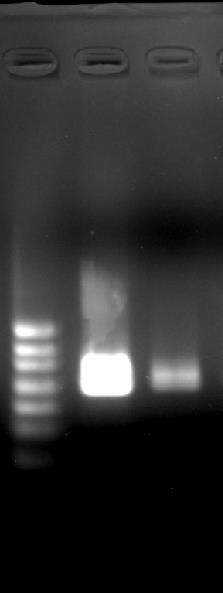
Figure 2. Agarose gel image of in vitro transcribed SARS-CoV-2 E gene RNA. Two 10-fold serial dilutions are loaded. Ladder sizes are corresponding to 1,000, 800, 600, 400, 300, and 200 bp from up.- Measure RNA concentration with QuantusTM Fluorometer.
- Calculate RNA copy number.
- Obtain molecular weight (M.W.) of the transcribed RNA (corresponding sequence: 5′-GGGGAAUUGUGAGCGGAUAACAAUUCCCCUCUAGAAAUAAUUUUGUUUAACUUUAAGAAGGAGAUAUACAUAUGUACUCAUUCGUUUCGGAAGAGACAGGUACGUUAAUAGUUAAUAGCGUACUUCUUUUUCUUGCUUUCGUGGUAUUCUUGCUAGUUACACUAGCCAUCCUUACUGCGCUUCGAUUGUGUGCGUACUGCUGCAAUAUUGUUAACGUGAGUCUUGUAAAACCUUCUUUUUACGUUUACUCUCGUGUUAAAAAUCUGAAUUCUUCUAGAGUUCCUGAUCUUCUGGUCUAACUCGAGCACCACCACCACCACCACUGAGAUCCGGCUGCUAACAAAGCCCGAAAGGAAGCUGAGUUGGCUGCUGCCACCGCUGAGCAAUAACUAGC-3′) using Polynucleotide Molecular Weight Calculator, or using following formula where “#N” represents the number of each nucleotide.
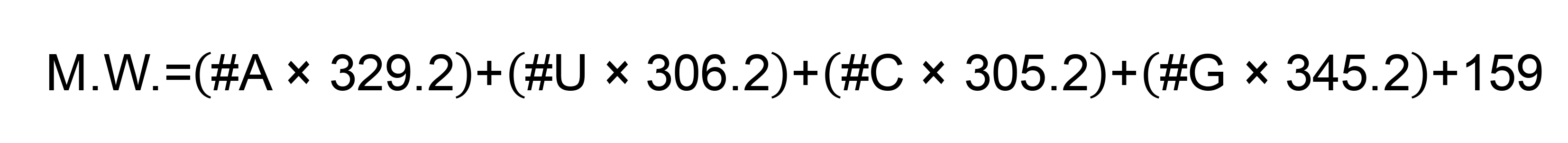
- Calculate copy number using following formula:
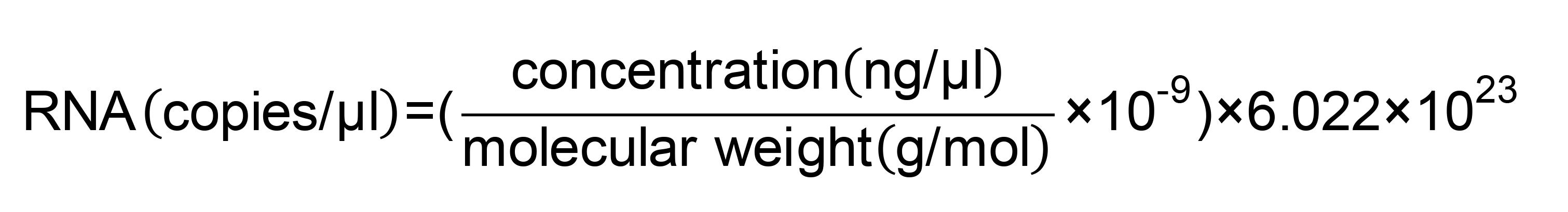
- Obtain molecular weight (M.W.) of the transcribed RNA (corresponding sequence: 5′-GGGGAAUUGUGAGCGGAUAACAAUUCCCCUCUAGAAAUAAUUUUGUUUAACUUUAAGAAGGAGAUAUACAUAUGUACUCAUUCGUUUCGGAAGAGACAGGUACGUUAAUAGUUAAUAGCGUACUUCUUUUUCUUGCUUUCGUGGUAUUCUUGCUAGUUACACUAGCCAUCCUUACUGCGCUUCGAUUGUGUGCGUACUGCUGCAAUAUUGUUAACGUGAGUCUUGUAAAACCUUCUUUUUACGUUUACUCUCGUGUUAAAAAUCUGAAUUCUUCUAGAGUUCCUGAUCUUCUGGUCUAACUCGAGCACCACCACCACCACCACUGAGAUCCGGCUGCUAACAAAGCCCGAAAGGAAGCUGAGUUGGCUGCUGCCACCGCUGAGCAAUAACUAGC-3′) using Polynucleotide Molecular Weight Calculator, or using following formula where “#N” represents the number of each nucleotide.
Make 10-fold serial dilutions in TE buffer as RT-qPCR standards.
Viral RNA Preparation and Titration Using E gene
Extract viral RNA from debris-free culture media with QIAamp Viral RNA Mini Kit following manufacturer’s instructions.
Aliquot viral RNA extract and store at -80 °C until use.
Titrate cultured viral RNA copy number by RT-qPCR
Prepare primer mix as follows: for 100 μl, mix 10 μl of forward and reverse primer, 5 μl of probe, 75 μl of DEPC treated water. Stock concentration of each primer/probe is 100 μM. Names and sequences of primers are listed in Table 1. Primer mix may be prepared in a batch and stored in -20 °C for later use.
Prepare master mix except template as follow : 7.5 μl of Luna® Universal Probe One-Step Reaction Mix, 0.75 μl of Luna® WarmStart® RT Enzyme Mix, 0.6 μl of primer mix, and 4.15 μl of DEPC treated water per reaction.
Aliquot 13 μl of master mix to each well of PCR tube or plate.
Add 2 μl of template (viral RNA dilutions, in vitro transcribed RNA standards or no template control) to each well. Close lid of PCR tube or cover film on plate.
Run PCR reaction as following temperature and time setting: 10 min at 55 °C, 1 min at 95 °C, and 45 cycles of 10 s at 95 °C and 30 s at 60 °C.
Copy number of viral RNA can be calculated by substituting threshold cycle value (Ct) to the equation of standard curve of following form:

Standard curve generation and sample copy number calculation can be done using each software coupled with real-time PCR instrument.
RT-LAMP reaction
Template viral RNA may prepared from cultured virus or biological specimen from COVID-19 patients. Selection of proper RNA extraction method is up to performers.
Prepare 10x LAMP primer mix as follows: for 100 μl, mix 16 μl of FIP/BIP, 2 μl of F3/B3 primers, 4 μl of LF/LB primers and 56 μl of DEPC treated water. Stock concentration of each primers is 100 μM. Sequences of primers are listed in Table 1. The LAMP primer mix may be prepared in a batch and stored in -20 °C for later use.
Prepare RT-LAMP reaction mix. Master mix except template can be prepared in a batch.
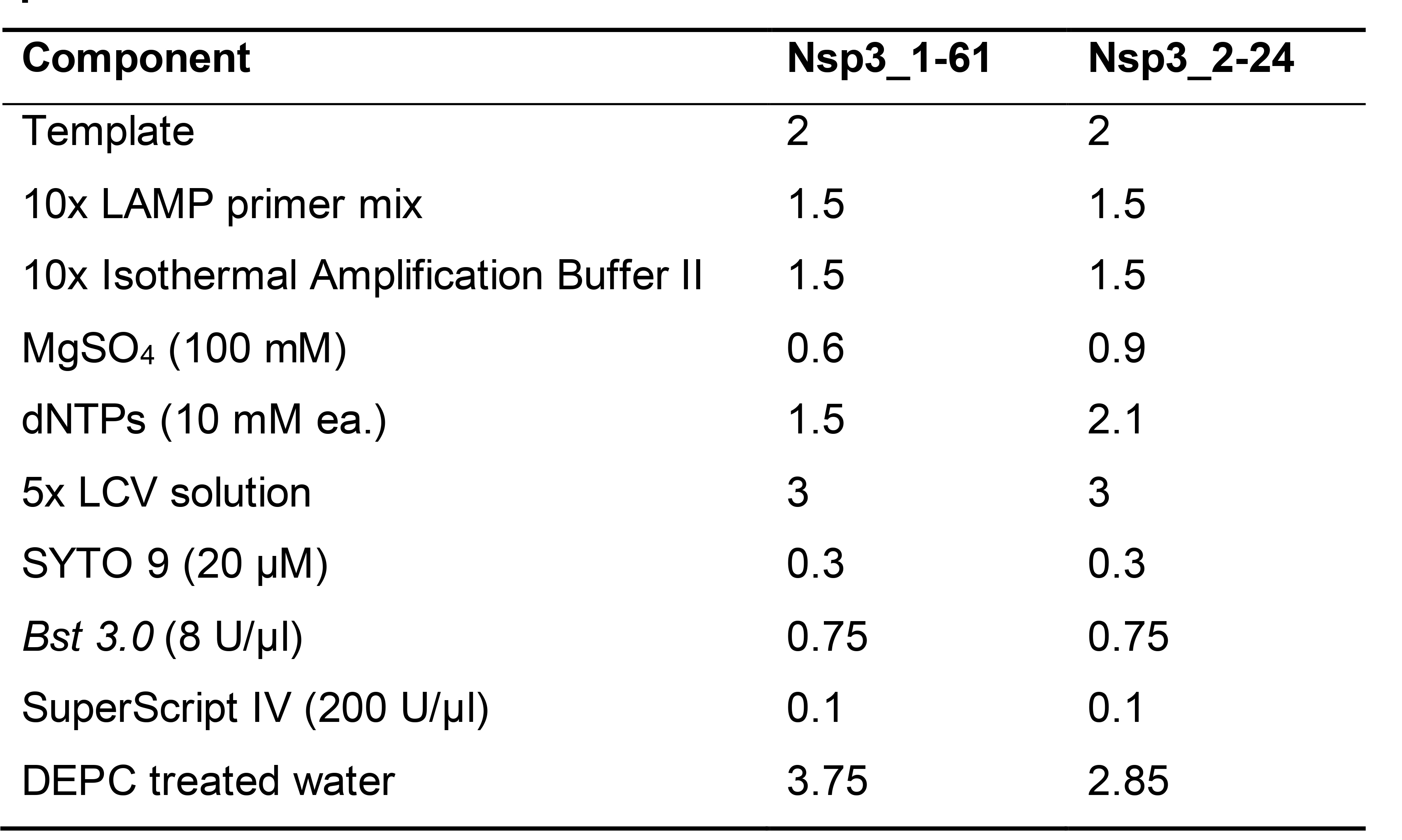
For LCV method, Reaction components, stock concentration and volume to use are as listed in Table 2.
When using WarmStart® Colorimetric LAMP 2X Master Mix, prepare reaction mix as follows: 2 μl of template, 1.5 μl of 10x LAMP primer mix, 0.3 μl of 20 μM SYTO9, 7.5 μl of LAMP mix, and 3.7 μl of DEPC treated water.
Table 2. Components and volume used (μl) for RT-LAMP with designated LAMP primer sets
Aliquot 13 μl of reaction mix and 2 μl of template to each well of PCR tube or plate and close lid or cover film.
Run isothermal amplification. Following example settings are for the LightCycler® 96 instrument SYTO 9 as fluorescent dye.
Select SYBR Green I and set “Integration Time” to 1 s.
Set cycling condition to 59 s at 65 °C for Nsp3_1-61 primer set or 69 °C for Nsp3_2-24 primer set. Use 65 °C for reaction with WarmStart® Colorimetric LAMP 2x Master Mix. Set 59 s of incubation time and 1 s of data acquisition time for each cycle to make actual incubation time per cycle as 1 min. Set number of cycles to the desired incubation time in minutes (Figure 3).
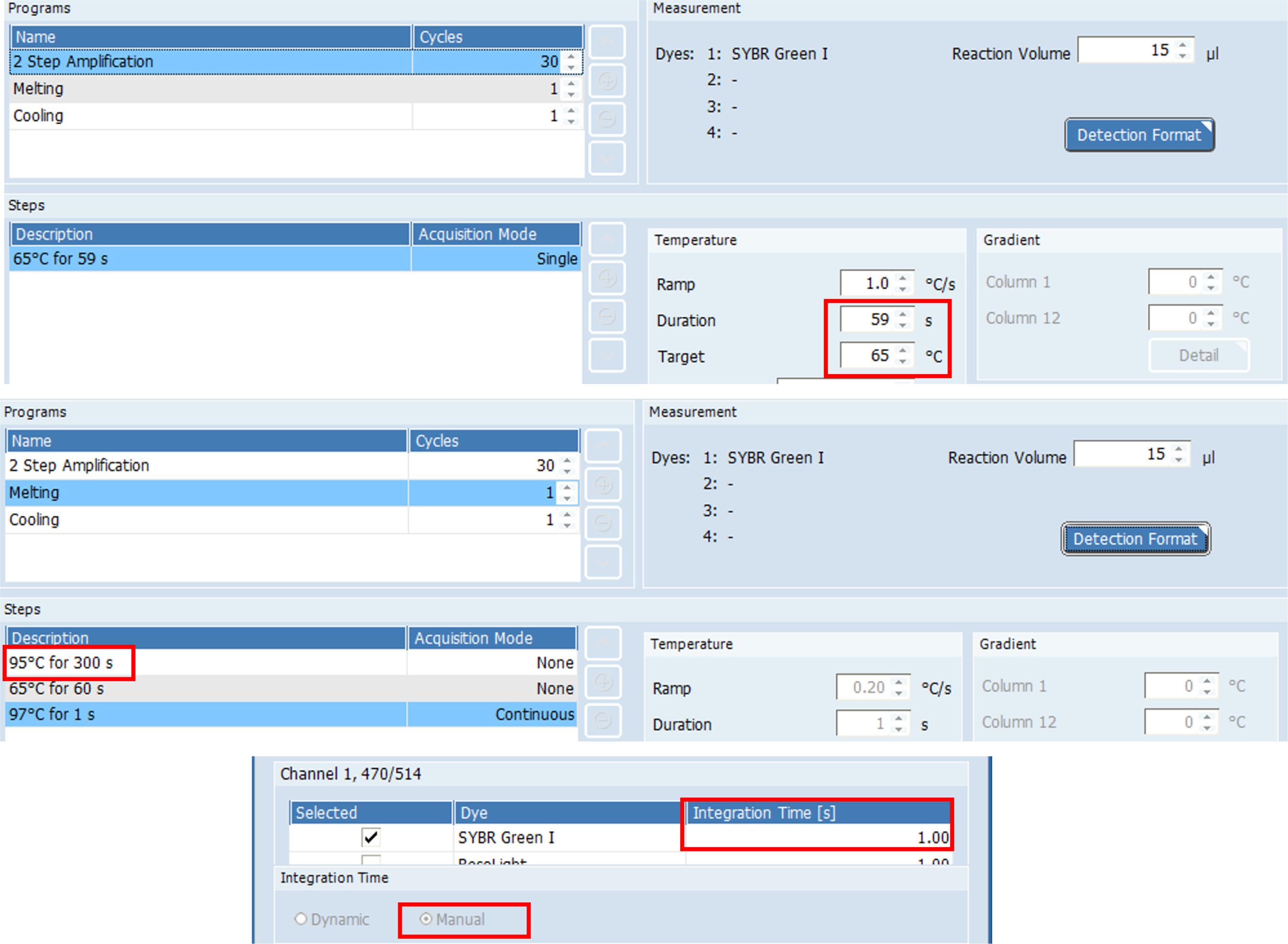
Figure 3. Example settings for the primer set Nsp3_1-61 using LightCycler® 96 software
Data analysis
Real-time amplification of LAMP reactions can be observed via LightCycler 96 software or other program accompanied with each qPCR machines. Sigmoidal amplification curve is shown for samples with positive result while no amplification is shown for negative samples or samples with negative results. Non-specific amplification curve may be observed with significantly delayed threshold time and/or shifted melting temperature (Figure 4).
For tests for which WarmStart Colorimetric LAMP 2x Master Mix is used, samples with positive result show yellow color while samples with negative result show pink color. For tests to which LCV method is adapted, samples with positive result show clear blue color from blueish transparent color (Figure 5).
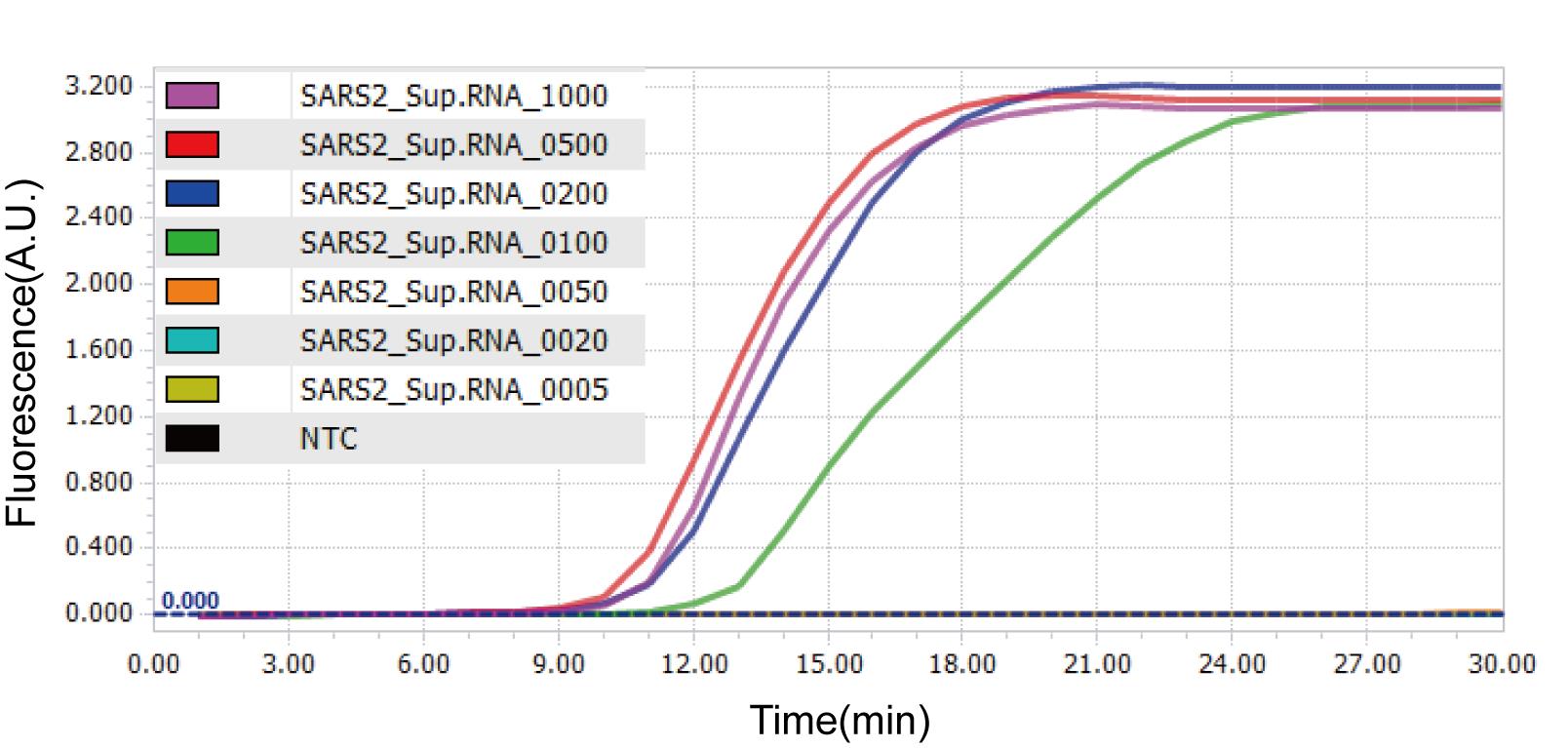
Figure 4. Amplification curves of RT-LAMP using Nsp3_1-61 primer set on LightCycler 96 system. Representative amplification curves of one replicate sample with designated RNA copies are shown. NTC, No Template Control.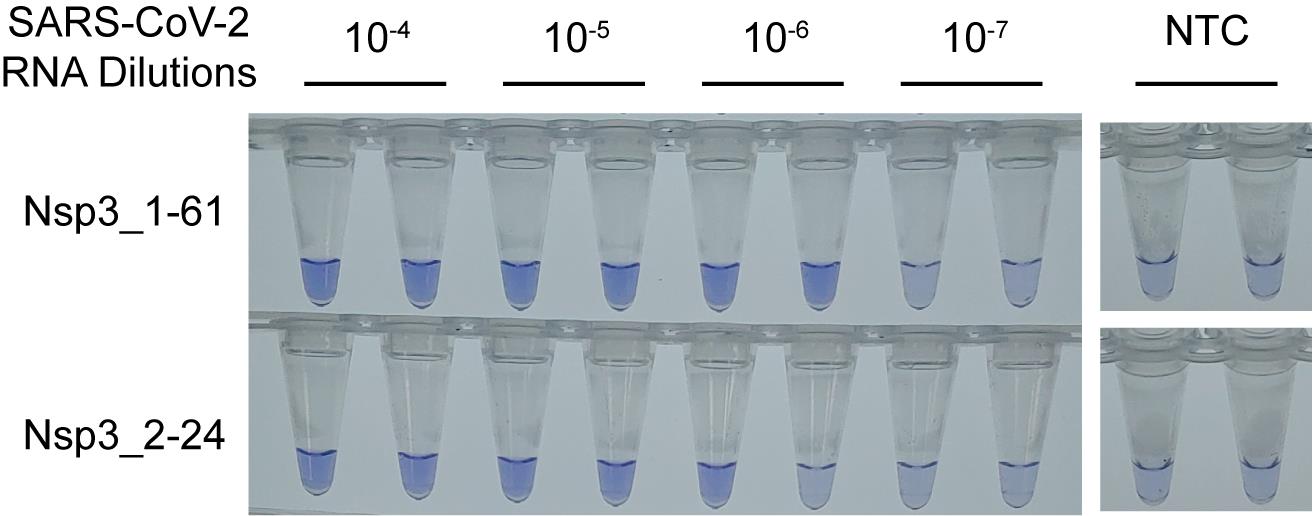
Figure 5. The color changes of LCV method for RT-LAMP. LCV color changes to blue by positive RT-LAMP amplification. RT-LAMP reaction was performed for 60 min at 69 °C with the half amount of SuperScript IV reverse transcriptase described in the main protocol as the result is from an experiment during reaction optimization. NTC, No Template Control.Notes
Here we targeted E gene for RT-qPCR titration while LAMP primer targets Nps3. Therefore titrated RNA copy number does not perfectly match with LAMP target copy number.
Denaturing agarose gel for RNA electrophoresis should be prepared in fume hood as vapor with formaldehyde is toxic. It would be better to do electrophoresis in fume hood too.
In vitro transcription usually give micrograms of RNA products.
Same as or more than seven points with 5 to 10 fold serial dilution are generally recommended for standards of RT-qPCR. We used 108 to 100 copies per reaction and no template control (NTC) for standard curve generation. Triplicate for each dilution is sufficient.
While we obtained about 107 copies/μl concentration from cultured virus, the number may differ by incubation period and virus species. Non-infected cells work as the negative control for viral infection yet the derived RNA is not necessarily included for RT-qPCR as no template control can serve as the negative control of RT-qPCR.
When preparing a master mix for RT-qPCR and LAMP/RT-LAMP in bulk for a batch of experiment, add extra volume to prevent shortage of the master mix from pipette error or other reasons. For 15 μl reaction described in this protocol, 0.5 to 1 extra amount for < 20 reactions or about 10% extra amount for more reactions is usually enough.
For RT-qPCR and real-time RT-LAMP, other qPCR instruments than LightCycler® 96 Software can be used. For real-time RT-LAMP, the time required for measuring fluorescence should be accounted for setting incubation time and cycle scheme and for the calculation between Ct and threshold time. For example, Bio-Rad CFX systems need rather unpredictable data acquisition time as the system’s detection unit scan each well one by one.
A melting step may be added to the RT-LAMP reaction since non-specific amplifications may be distinguished from their melting curves. Addition of an incubation step for 5 min at 95 °C between amplification and melting steps is rather recommended for polymerase inactivation.
When using LCV method for colorimetric detection, color varies by temperature. In high temperature, color of reaction mixture may turn to blue without nucleic acid amplification. Color of the mixture will back to normal in room temperature (18-25 °C) within 5 min.
The sensitivity of LAMP is bottlenecked by formation of ‘dumbbell intermediate’ and it is usually lower than that of PCR. However, LAMP is highly vulnerable to cross contamination because LAMP yield is high and the bottleneck is not applied. Therefore, we strongly discourage to open reaction tube after LAMP reaction. Otherwise, using separated space for analysis of reaction product (e.g., agarose gel electrophoresis) is recommended.
Typical LAMP optimization includes changing concentration of dNTPs or Mg2+. We suggest to test each concentration in combination because dNTPs can chelate Mg2+.
Since EDTA in TE buffer chelate Mg2+ ion, using DEPC treated water for RNA elution and dilution may be beneficial when testing larger volume of template.
Recipes
Denaturing agarose gel (1%, 100 ml)
1g agarose
10 ml 10x MOPS
18 ml 37% formaldehyde
72 ml DEPC treated water
20 μM SYTO 9
Dissolve 5 mM SYTO 9 original stock in DEPC treated water with 1/250 dilution. Aliquot in small volume (~100 μl) and store at -20 °C
5x LCV solution
10.20 mg Crystal Violet
378.12 mg Sodium Sulfite
283.75 mg β-Cyclodextrin
Dissolved in 50 ml DEPC treated water
Acknowledgments
Authors thank to Korea Centers for Disease Control and Prevention (KCDC) for kind and rapid sharing of isolated strain of SARS-CoV-2. This work was supported by the National Research Council of Science and Technology (NST) grant by the Ministry of Science and ICT (Grant No. CRC-16-01-KRICT). The protocol provided is from our original article previously published ( Park et al., 2020 ).
Competing interests
The authors declare no conflict of interests.
References
- Corman, V. M., Landt, O., Kaiser, M., Molenkamp, R., Meijer, A., Chu, D. K., Bleicker, T., Brunink, S., Schneider, J., Schmidt, M. L., Mulders, D. G., Haagmans, B. L., van der Veer, B., S., den Brink, Wijsman, L., Goderski, G., Romette, J. L., Ellis, J., Zambon, M., Peiris, M., Goossens, H., Reusken, C., Koopmans, M. P. and Drosten, C. (2020). Detection of 2019 novel coronavirus(2019-nCoV) by real-time RT-PCR. Euro Surveill 25(3). doi: 10.2807/1560-7917.ES.2020.25.3.2000045..
- Goto, M., Honda, E., Ogura, A., Nomoto, A. and Hanaki, K. (2009). Colorimetric detection of loop-mediated isothermal amplification reaction by using hydroxy naphthol blue. Biotechniques 46(3): 167-172.
- Lalli, M. A., Chen, X., Langmade, S. J., Fronick, C. C., Sawyer, C. S., Burcea, L. C., Fulton, R. S., Heinz, M., Buchser, W. J., Head, R. D., Mitra, R. D. and Milbrandt, J. (2020). Rapid and extraction-free detection of SARS-CoV-2 from saliva with colorimetric LAMP. medRxiv .
- Miyamoto, S., Sano, S., Takahashi, K. and Jikihara, T. (2015). Method for colorimetric detection of double-stranded nucleic acid using leuco triphenylmethane dyes. Anal Biochem 47328-33.
- Niemz, A., Ferguson, T. M. and Boyle, D. S. (2011). Point-of-care nucleic acid testing for infectious diseases. Trends Biotechnol 29(5): 240-250.
- Notomi, T., Okayama, H., Masubuchi, H., Yonekawa, T., Watanabe, K., Amino, N. and Hase, T. (2000). Loop-mediated isothermal amplification of DNA. Nucleic Acids Res 28(12): E63.
- Oscorbin, I. P., Belousova, E. A., Zakabunin, A. I., Boyarskikh, U. A. and Filipenko, M. L. (2016). Comparison of fluorescent intercalating dyes for quantitative loop-mediated isothermal amplification(qLAMP). Biotechniques 61(1): 20-25.
- Park, G. S., Ku, K., Baek, S. H., Kim, S. J., Kim, S. I., Kim, B. T. and Maeng, J. S. (2020). Development of Reverse Transcription Loop-Mediated Isothermal Amplification Assays Targeting Severe Acute Respiratory Syndrome Coronavirus 2(SARS-CoV). J Mol Diagn 22(6): 729-735.
- Tanner, N. A., Zhang, Y. and Evans, T. C. (2015). Visual detection of isothermal nucleic acid amplification using pH-sensitive dyes. Biotechniques 58(2): 59-68.
- WHO. (2020). Coronavirus disease(COVID-19) technical guidance: Laboratory testing for 2019-nCoV in humans. Retrieved from https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/laboratory-guidance.
- Yan, C., Cui, J., Huang, L., Du, B., Chen, L., Xue, G., Li, S., Zhang, W., Zhao, L., Sun, Y., Yao, H., Li, N., Zhao, H., Feng, Y., Liu, S., Zhang, Q., Liu, D. and Yuan, J. (2020). Rapid and visual detection of 2019 novel coronavirus(SARS-CoV-2) by a reverse transcription loop-mediated isothermal amplification assay. Clin Microbiol Infect 26(6): 773-779.
- Zhang, Y., Odiwuor, N., Xiong, J., Sun, L., Nyaruaba, R. O., Wei, H. and Tanner, N. A. (2020). Rapid Molecular Detection of SARS-CoV-2(COVID-19) Virus RNA Using Colorimetric LAMP. medRxiv .
Article Information
Copyright
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Park, G., Baek, S., Ku, K., Kim, S. J., Kim, S. I., Kim, B. and Maeng, J. (2020). Colorimetric RT-LAMP Methods to Detect Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). Bio-protocol 10(21): e3804. DOI: 10.21769/BioProtoc.3804.
Category
Microbiology > Pathogen detection > RT-LAMP
Molecular Biology > RNA > RNA detection
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









