- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Derivation of Induced Pluripotent Stem Cells from Human Fibroblasts Using a Non-integrative System in Feeder-free Conditions
Published: Vol 10, Iss 20, Oct 20, 2020 DOI: 10.21769/BioProtoc.3788 Views: 5501
Reviewed by: Chiara AmbrogioJulie Sandra Di MartinoAna Santos Almeida

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Isolation and ex vivo Expansion of Human Limbal Epithelial Progenitor Cells
Naresh Polisetti [...] Ursula Schlötzer-Schrehardt
Sep 20, 2020 5104 Views

CRISPR/Cas9-mediated Precise SNP Editing in Human iPSC Lines
Hanwen Zhang and Siwei Zhang
Jun 20, 2021 6351 Views

Reprogramming Cancer Cells to Antigen-presenting Cells
Alexandra G. Ferreira [...] Carlos-Filipe Pereira
Nov 20, 2023 5033 Views
Abstract
Induced pluripotent stem cells (iPSCs) are genetically reprogrammed somatic cells that exhibit features identical to those of embryonic stem cells (ESCs). Multiple approaches are available to derive iPSCs, among which the Sendai virus is the most effective at reprogramming different cell types. Here we describe a rapid, efficient, safe, and reliable approach to reprogram human fibroblasts into iPSCs that are compatible with future iPSCs uses such as genome editing and differentiation to a transplantable cell type.
Keywords: Feeder-freeBackground
Induced pluripotent stem cells (iPSCs) are genetically reprogrammed adult cells that exhibit morphological and functional qualities remarkably similar to those of embryonic stem cells (ESCs) (Takahashi and Yamanaka, 2006; Yu et al., 2007). They offer a great opportunity not only for disease modeling, but for the development of therapeutic strategies for pathologies that involve tissue degeneration. Furthermore, the iPSCs promise relies on a safe replenishable cell source derived in chemically defined media and free of random DNA integration.
Reprogramming somatic cells into iPSCs requires the forced expression of transcription factors that support the pluripotent state, including OCT4, SOX2, KLF4, c-MYC, NANOG, and LIN-28 (Takahashi and Yamanaka, 2006; Takahashi et al., 2007; Yu et al., 2007). Multiple approaches are available to deliver the transcription factors into the cells, including those that require integration into the host chromosomes (Takahashi and Yamanaka, 2006; Kane et al., 2010). Exogenous DNA integration can lead to unpredictable effects on the quality of the cells and safety after transplantation. Other approaches include DNA based vectors that exist episomally (Yu et al., 2011; Weltner et al., 2012) and thus, decrease the possibility of integration, and finally those that do not integrate into the host genome and are known as transgene-free. The transgene-free methods include mRNA (Warren and Wang, 2013), recombinant protein (Zhou et al., 2009) and Sendai virus (Fusaki et al., 2009). Delivering the pluripotency transcription factors as mRNA or recombinant protein is poorly effective and costly. In contrast, Sendai virus, is a highly effective RNA virus that efficiently reprogram different types of somatic cells.
Sendai is a cytoplasmic RNA replication incompetent virus (SeV) that safely and effectively delivers the reprogramming factors into somatic cells, and does not integrate into the genome or alter the genetic information of the cell (Li et al., 2000; Fusaki et al., 2009). Furthermore, the virus is cleared out from the cells after a few passages assuring zero footprint of both the vectors and transgenes. The Sendai reprogramming system is commercially available as CytoTune iPS 2.0 for research purposes and as CTS CytoTune iPS 2.1 for clinical use. Both systems contain vectors encoding the four Yamanaka factors (OCT4, SOX2, KLF4, and c-MYC) optimized for generating iPSCs from human somatic cells, making Sendai virus the most rapid, efficient and cost-efficient method to generate transgene-free iPSCs.
Reliable and safe derivation of human iPSCs relies on the use of defined and qualified reagents that allow a smooth transition to downstream technologies and is compatible with GMP (Good Manufacture Practice) quality standards for large-scale cell production. Here we describe a reprogramming approach for human fibroblasts that uses a non-integrative system, chemically defined culture medium in feeder-free conditions. This approach enables rapid, efficient, safe and reliable derivation of iPSCs compatible with future uses including genome editing and differentiation to a transplantable cell type.
Materials and Reagents
- Falcon® 6-Well Flat-Bottom Plate, Tissue Culture-Treated (Falcon, catalog number: 38016 )
- Serological pipettes multiple sizes (Corning)
- Falcon 15 ml conical centrifuge tubes (Falcon, catalog number: 352097 )
- Falcon 50 ml conical centrifuge tubes (Falcon, catalog number: 352070 )
- Cells: Human fibroblast expanded from skin biopsies preferentially at passage 1 to passage 5. Early passage fibroblast can be obtained from a commercial source (i.e., ATCC), a repository (i.e., Corriell Institute) or expanded in the lab from skin biopsies (Vangipuram et al., 2013 describes the procedure in detail)
- Corning® Matrigel® hESC-Qualified Matrix (Corning, catalog number: 354277 )
- CytoTuneTM-iPS 2.0 Sendai Reprogramming Kit (Thermo Scientific, catalog number: A16518 )
- DMEM/F-12 (Gibco, catalog number: 11320033 )
- Fetal Bovine Serum (Gibco, catalog number: 16000044 )
- MEM NEAA (Gibco, catalog number: 11140050 )
- Pen Strep (Gibco, catalog number: 15-140-122 )
- mTeSRTM 1 (STEMCELL Technologies, catalog number: 85850 )
- TrypLETM Express Enzyme (1x) (Gibco, catalog number: 12605036 )
- Anti-Human TRA-1-60, Mouse monoclonal (Thermo Fisher Scientific, catalog number: 41-1000 )
- Anti-Human OCT4, Rabbit polyclonal, IgG1 (Abcam, catalog number: ab19857 )
- Anti-Human SOX2 Antibody, Rabbit polyclonal IgG1 (Abcam, catalog number: ab97959 )
- STEMdiff Trilineage Differentiation kit, STEMCELL Technologies, catalog number: 0 5230 )
- Universal Mycoplasma Detection Kit (ATCC, catalog number: 30-1012K )
- Dulbecco’s PBS (DPBS) without Calcium and Magnesium (Gibco, catalog number: 14190136 )
- Polybrene Hexadimethrine Bromide (Sigma-Aldrich, catalog number: H9268 )
- 0.5 M EDTA (Gibco, catalog number: 15575020 )
- Fibroblast culture medium (see Recipes)
- 5 mM EDTA solution (see Recipes)
- 10 mM Y27632 stock solution (see Recipes)
Equipment
- Tissue culture incubator HeracellTM 150i (Thermo Fisher Scientific, catalog number: 51026283 )
- Beckman Avanti J-30I Refrigerated with plate adapters (Beckman Coulter, catalog number: 17039 )
- Tissue culture laminar flow hood (NuAire Class II Type A2 NU-540)
- Evos XL core microscope (Thermo Scientific, catalog number: AMEX1100 )
- Countess II FL Automated Cell Counter (Thermo Fisher Scientific, catalog number: AMQAF1000 )
- Water bath Isotemp (Fisher Scientific, catalog number: S35936 )
- Portable Pipet-aid (Drummond, model: DP-101 , catalog number: 4-000-101)
- Micro pipettes (Eppendorf)
Procedure
Day -1: Plate dermal fibroblast for reprogramming
Use low-passage (passage 1 to 5) human fibroblasts for reprogramming experiments. The reprogramming efficiency decreases with each passage. Hence, we don’t recommend reprogramming fibroblast after passage 6. A schematic timeline and representative pictures can be seen in Figure 1.
- Coat two wells of a 6 well plate with 2 ml of 5 mg/ml Matrigel and make sure it covers the entire well. Incubate at 37 °C for 30 min.
- Inspect the fibroblast culture for the desired confluency (more than 70%), aspirate the culture medium and rinse twice with 2 ml DPBS.
- Add 500 µl TrypLETM Express enzyme and incubate at 37 °C and 5% CO2 for 2 to 5 min or until fibroblasts have detached. TrypLETM is used because it has lower cell toxicity than standard Trypsin and it is an animal-free product.
- Add 1 ml fibroblast culture medium and transfer cell suspension to a 15-ml conical tube.
- Centrifuge at 200 x g for 5 min. Remove and discard supernatant.
- Resuspend cells in 1 ml of fresh fibroblast culture medium. Recipe available in the recipe section.
- Count fibroblasts using the desired method (e.g., Countess Automated Cell Counter), and plate 1 x 105 cells/well (10,000 cells/cm2) in the Matrigel-coated 6-well dish in 2 ml of fibroblast culture medium.
- Incubate 24 h at 37 °C with a humidified atmosphere and 5% CO2.
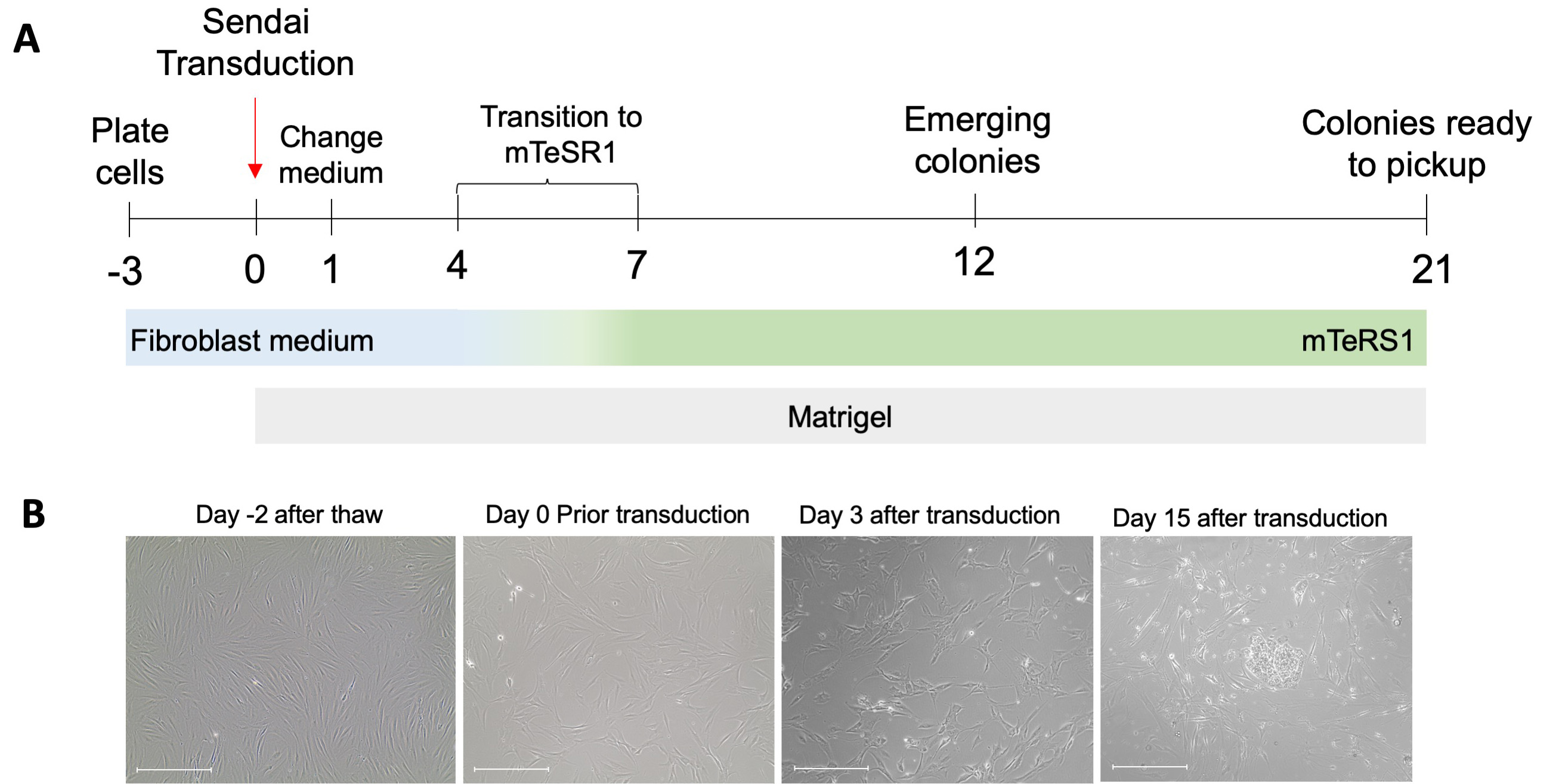
Figure 1. Derivation of induced pluripotent stem cells from fibroblast using Sendai virus and chemically defined medium. A. Schematic view of general reprogramming procedure including culture medium used. B. Images of fibroblast morphology changes during the early stages of reprogramming. Scale bars: 650 μm.
- Inspect the fibroblast culture, proceed with transduction if cell density is approximately 40-50% confluent, and cells have fully adhered and extended. If those parameters are not meet, wait another 24 h.
- Warm 1 ml of fibroblast culture medium in a water bath for each well to be transduced calculate the volume of each premade virus needed to reach a multiplicity of infection of (MOI) of 5:5:3 (KOS MOI = 5, hc-Myc MOI = 5, hKlf4 MOI = 3) using the live cell count from the day of the seeding, and the titer information on the CoA (calculate based on the lot number, the CoA can be download from thermofisher.com/cytotune). Calculate the volume of virus using the formula:
CIU = Cell infectious units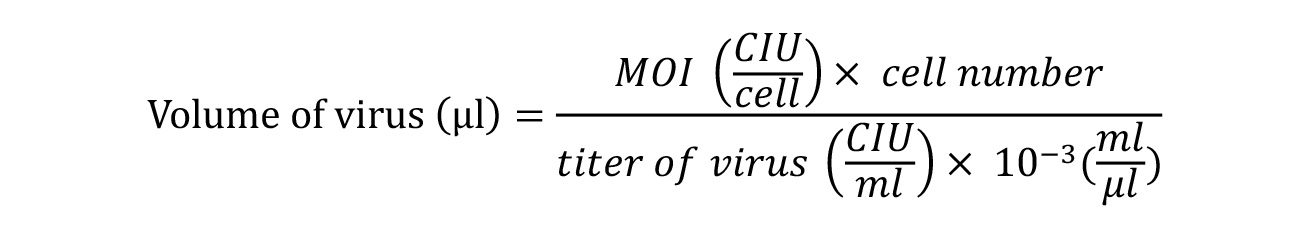
For example, to calculate the volume of hKOS virus with a titter of 1.1 x 108 CIU;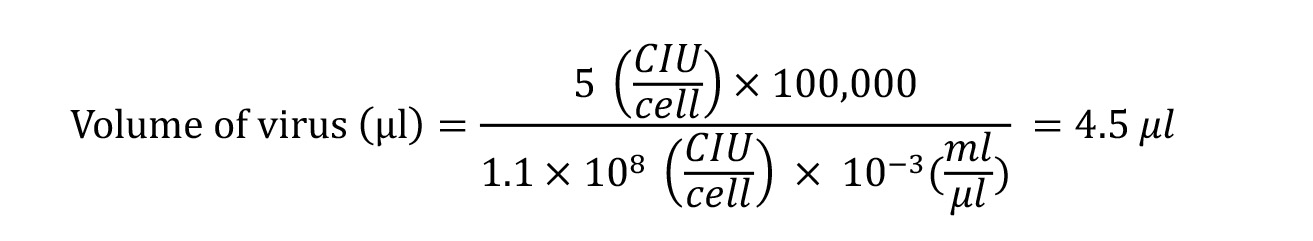
- Thaw one set of CytoTuneTM 2.0 Sendai aliquots from -80 °C and briefly centrifuge the tube. Place it immediately on ice until ready to use.
- Add the calculated volumes of each of the three CytoTuneTM 2.0 Sendai virus to 1 ml of pre-warmed fibroblast culture medium.
- OPTIONAL: Add 4 μg/ml of polybrene to the medium. Ensure that the solution of virus and polybrene is thoroughly mixed by pipetting the mixture gently up and down. Complete the next step within 5 min.Polybrene increases the transduction efficiency of the virus, however, it can be toxic for some cell types. It neutralizes the charge repulsion between the virus and the cell surface increasing the overall transduction efficiency.
- Aspirate the fibroblast culture medium from the cells, and add the reprogramming virus and polybrene mixture to the well containing the cells.
- Close the 6-well dish, securely place on the plate adaptor, and centrifuge at 1,200 x g for 45 min at room temperature. Once the centrifugation is complete, add an additional 1 ml of prewarm fibroblast culture medium.
- Incubate the plate overnight in a 37 °C incubator with a humidified atmosphere of 5% CO2.
After 24 h, aspirate medium with viruses and add 2 ml fresh fibroblast culture medium. Expect to see cytotoxicity 24-48 h post-transduction, this is an indication of high uptake of the virus. See Figure 2.
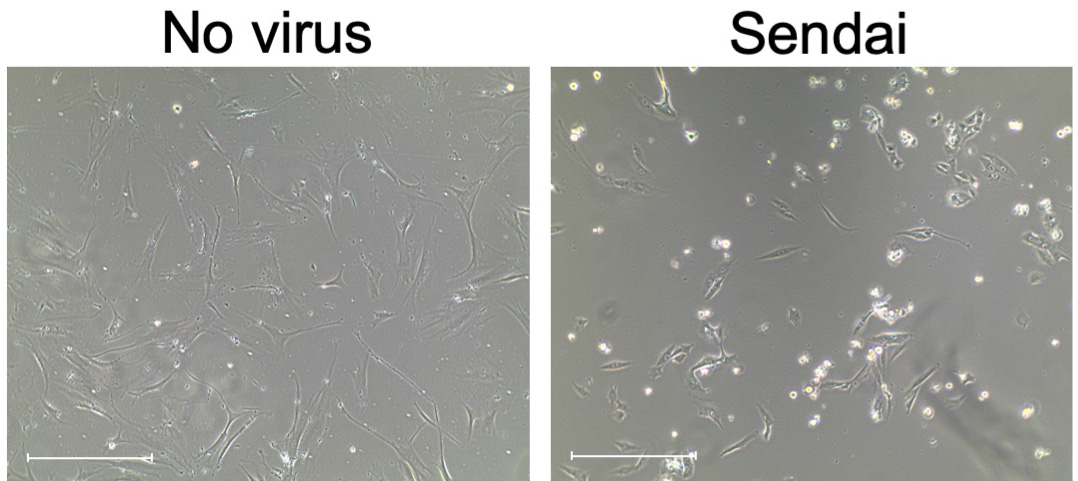
Figure 2. Images of morphological changes seventy-two hours after transduction with Sendai virus. Scale bars: 650 μm.
Days 2 to 3
Remove 2 ml of used medium and add 2 ml of fibroblast culture medium. Expect to see changes in cell morphology.
Day 4: Transition to defined medium
There are many different chemically defined culture medium commercially available for deriving and maintaining iPSCs. We have used mTeSR1 medium (Stem Cell Technologies), STEMFLEX (Life technologies) and mTeSR plus (Stem Cell Technologies) agnostically.
- Remove 500 μl of fibroblast culture medium and add 500 μl of fresh mTeSR1.
Day 5
- Remove 1 ml of culture medium and add 1 ml of fresh mTeSR1.
Day 6
- Remove 1.5 ml of culture medium and add 1.5 ml of fresh mTeSR1.
Day 7
Remove 2 ml of medium culture medium and add the same volume of fresh mTeSR1.
Days 8-20
Feed cells daily with 2 ml until colonies are ready to be passaged. Remove partially reprogrammed and differentiated colonies by scraping them before medium change. Fully reprogrammed colonies have round shape with well-defined borders, cells display identical morphology with a high ratio of nucleus to cytoplasm and prominent nucleoli. In contrast, partially reprogrammed colonies have undefined borders, an amorphous shape, and are composed of different types of cells.
Day 21: First passage
- When colonies are ready to pick, prepare one Matrigel coated dish per colony to be passage, and incubate at 37 °C for at least 30 min.
- Aspirate Matrigel from plates and add 2 ml mTeSR1 + 10 µM ROCK inhibitor.
- Use a 22 gauge needle or a pulled glass pipette to cut colonies in a grid-like pattern into small fragments. See Figure 3.
- Use a 100 µl pipettor to scrape fragments and collect them. Immediately transfer the fragments to the Matrigel coated plates with mTeSR1 + 10 µM ROCK inhibitor.
- Rock plate back-and-forth and side-to-side to evenly distribute the cell fragments and incubate overnight at 37 °C with a humidified atmosphere of 5% CO2.
- Feed cells daily until ready to passage (usually every 6 to 7 days).
- Repeat Steps 1 to 10 for 7 passages. We found that > 95% of the iPSCs clones at passage 7, are transgene free.
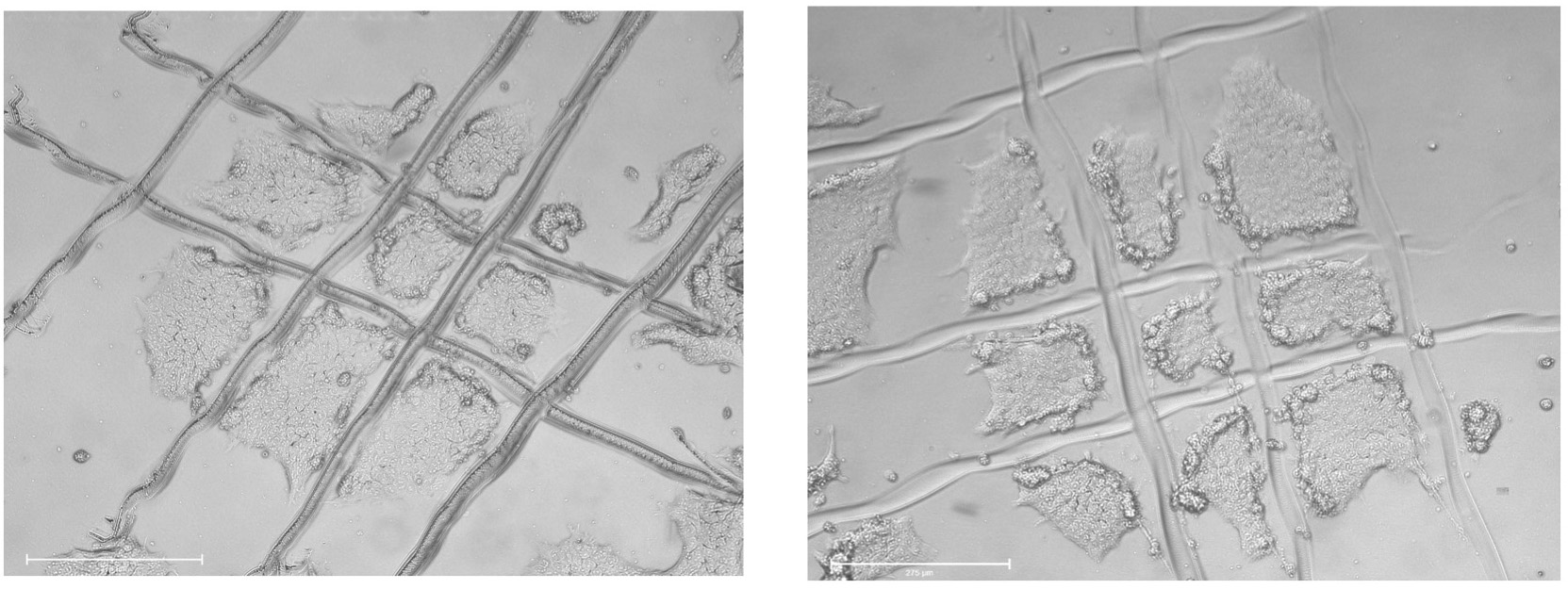
Figure 3. Images of colonies cut in a grid-like pattern into small fragments. Scale bars: 650 μm.
After passage 7, IPSCs are passage with EDTA buffer, as follow:
- Coat 6 wells of a 6-well tissue culture plate with Matrigel and place in incubate at 37 °C for 30 min.
- Prior to use, allow the Matrigel-coated plate to equilibrate to room temperature for at least 1 h.
- Just before dissociating cells for passaging, aspirate the liquid Matrigel solution from the wells and replace with 1 ml of mTeSR1 + 10mM Y27632 cell culture media per well. Set aside.
- Wash the cells with 2 ml 0.5 mM EDTA, aspirate.
- Add 2 ml of room temperature 0.5 mM EDTA solution to the cells.
- Incubate the culture at 37 °C for 3 to 5 min, or until cells begin to separate uniformly throughout the entire colony. Do not allow the cultures detach in the EDTA solution.
- As soon as the cells appear rounded and uniform separation is seen throughout the colonies, carefully aspirate the EDTA solution from the well. Do not rinse.
- Immediately add 1 ml of mTeSR1 + 10mM Y27632. With a 5 ml pipet, take up the 1 ml of media from the well, and very gently dispense it against the culture surface to dissociate the cells from the dish. Repeat 1 to 2 more times, if needed.
- Be careful not to over-pipet the cell suspension.
- Dispense the cells gently into the 15 ml conical tube containing an additional 3 ml of pre-warmed media.
- Pipet the solution very gently 1 time to mix, and dispense 1 ml of the cell suspension drop-wise into each of the 6 new Matrigel-coated wells, and immediately rock plate back-and-forth and side-to-side to evenly distribute the colony pieces across the well.
- Incubate undisturbed at 37 °C and 5% CO2 overnight.
- Replace cell culture media every day with 2.5 ml of fresh mTeSR1, warmed to room temperature.
- Monitor cells daily and passage as needed.
Cells are expanded for cryopreservation at passage 10. Then, iPSCs are characterized (Figure 4) and prepare for downstream usage in genome engineering experiments or differentiation into specific cell types. Characterization includes:
- Immunofluorescence staining of cell surface markers such Anti-Human TRA-1-60, Mouse monoclonal and transcription factors such as anti-Human OCT4, Rabbit polyclonal, IgG1 and anti-Human SOX2 Antibody, Rabbit polyclonal IgG1.
- Pluripotency and trilineage differentiation capabilities (using the STEMdiff Trilineage Differentiation kit, and the Taqman hPSC Score card Panel.
- Test for mycoplasma contamination using commercially available kits such as The Universal Mycoplasma Detection Kit.
- G-banding Karyotype.
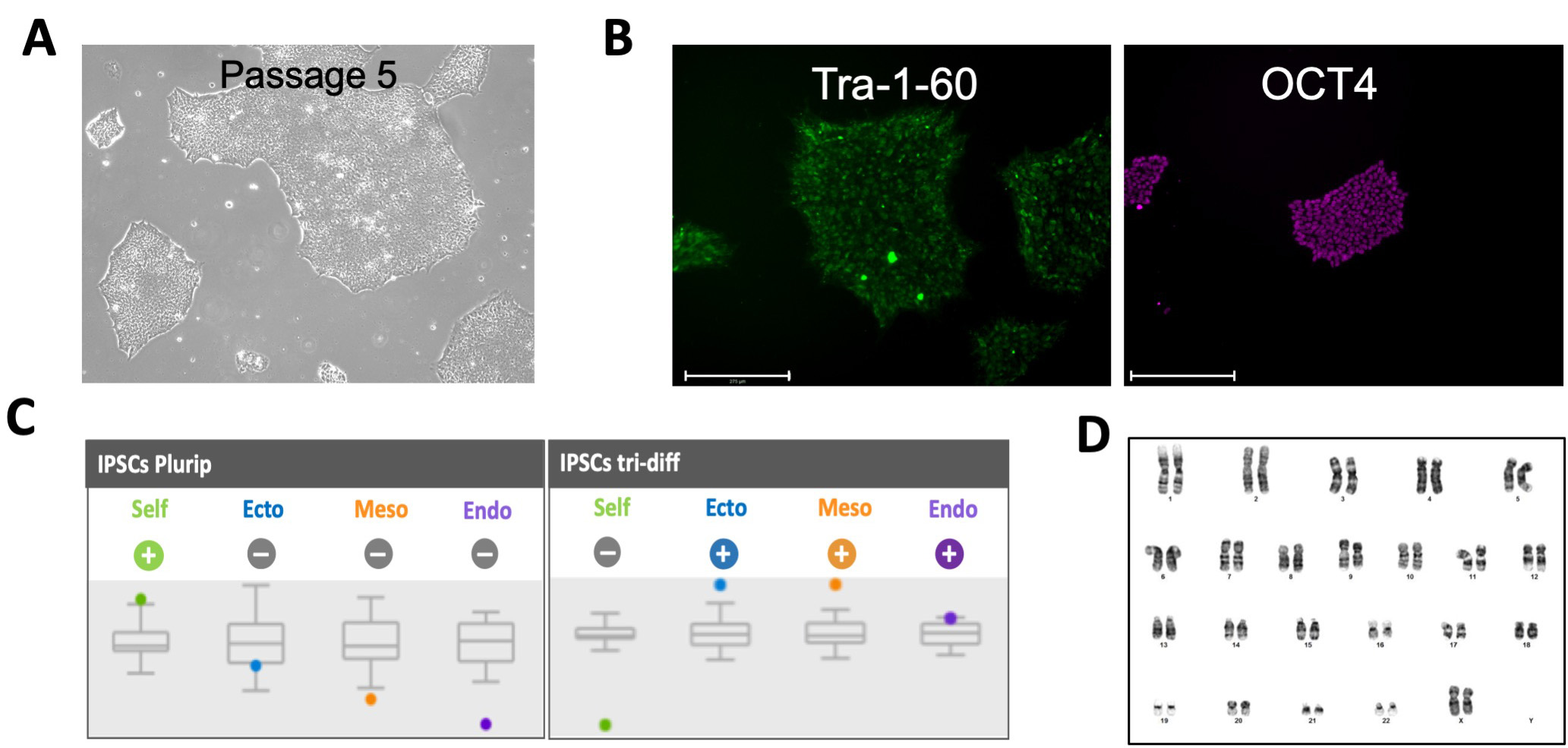
Figure 4. Characterization of iPSC derived from fibroblast. A. Bright field images of iPSCs, B. Immunofluorescence staining for pluripotency markers Tra-1-60 and OCT4. C. Taqman ScoreCard assay for pluripotent iPSCs (left panel) and iPSCs differentiated into endoderm, mesoderm and ectoderm (right panel). D. G-banding karyotype.
Recipes
- Fibroblast culture medium
The following recipe is to prepare 500 ml of fibroblast culture medium. Aseptically mix the following:
445 ml high glucose DMEM
50 ml heat-inactivated fetal bovine serum (FBS)
5 ml non-essential amino acids - 5 mM EDTA solution
Dilute 0.5 ml of 0.5 M EDTA in 500 ml DPBS without Calcium and Magnesium. Store at room temperature - 10 mM Y27632 stock solution
Aseptically add 3.122 ml of sterile water to 10 mg of Y27632
Mix thoroughly by pipetting, aliquot in Eppendorf’s and store at -80 °C
Avoid repeated freezing and thawing
Acknowledgments
This protocol was developed as part of standard operations procedures of the Human Pluripotent Stem Cell core facility at UNC-CH. This work is supported by the UNC Office of Research.
Competing interests
The authors do not have any financial or non-financial interest in the subject matter or materials discussed in this manuscript.
Ethics
This research was carried out in according with the UNC-CH institutional guidelines.
References
- Fusaki, N., Ban, H., Nishiyama, A., Saeki, K. and Hasegawa, M. (2009). Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proc Jpn Acad Ser B Phys Biol Sci 85(8): 348-362.
- Kane, N. M., Nowrouzi, A., Mukherjee, S., Blundell, M. P., Greig, J. A., Lee, W. K., Houslay, M. D., Milligan, G., Mountford, J. C., von Kalle, C., Schmidt, M., Thrasher, A. J. and Baker, A. H. (2010). Lentivirus-mediated reprogramming of somatic cells in the absence of transgenic transcription factors. Mol Ther 18(12): 2139-2145.
- Li, H. O., Zhu, Y. F., Asakawa, M., Kuma, H., Hirata, T., Ueda, Y., Lee, Y. S., Fukumura, M., Iida, A., Kato, A., Nagai, Y. and Hasegawa, M. (2000). A cytoplasmic RNA vector derived from nontransmissible Sendai virus with efficient gene transfer and expression. J Virol 74(14): 6564-6569.
- Takahashi, K., Tanabe, K., Ohnuki, M., Narita, M., Ichisaka, T., Tomoda, K. and Yamanaka, S. (2007). Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131(5): 861-872.
- Takahashi, K. and Yamanaka, S. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126(4): 663-676.
- Vangipuram, M., Ting, D., Kim, S., Diaz, R. and Schüle, B. (2013). Affiliations expand Skin punch biopsy explant culture for derivation of primary human fibroblasts. J Vis Exp (77): e3779.
- Warren, L. and Wang, J. (2013). Feeder-free reprogramming of human fibroblasts with messenger RNA. Curr Protoc Stem Cell Biol 27: Unit 4A 6.
- Weltner, J., Anisimov, A., Alitalo, K., Otonkoski, T. and Trokovic, R. (2012). Induced pluripotent stem cell clones reprogrammed via recombinant adeno-associated virus-mediated transduction contain integrated vector sequences. J Virol 86(8): 4463-4467.
- Yu, J., Chau, K. F., Vodyanik, M. A., Jiang, J. and Jiang, Y. (2011). Efficient feeder-free episomal reprogramming with small molecules. PLoS One 6(3): e17557.
- Yu, J., Vodyanik, M. A., Smuga-Otto, K., Antosiewicz-Bourget, J., Frane, J. L., Tian, S., Nie, J., Jonsdottir, G. A., Ruotti, V., Stewart, R., Slukvin, II and Thomson, J. A. (2007). Induced pluripotent stem cell lines derived from human somatic cells. Science 318(5858): 1917-1920.
- Zhou, H., Wu, S., Joo, J. Y., Zhu, S., Han, D. W., Lin, T., Trauger, S., Bien, G., Yao, S., Zhu, Y., Siuzdak, G., Scholer, H. R., Duan, L. and Ding, S. (2009). Generation of induced pluripotent stem cells using recombinant proteins. Cell Stem Cell 4(5): 381-384.
Article Information
Copyright
Beltran et al. This article is distributed under the terms of the Creative Commons Attribution License (CC BY 4.0).
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Beltran, A. A., Molina, S. G. and Beltran, A. (2020). Derivation of Induced Pluripotent Stem Cells from Human Fibroblasts Using a Non-integrative System in Feeder-free Conditions. Bio-protocol 10(20): e3788. DOI: 10.21769/BioProtoc.3788.
- Battaglia, R. A., Beltran, A. S., Delic, S., Dumitru, R., Robinson, J. A., Kabiraj, P., Herring, L. E., Madden, V. J., Ravinder, N., Willems, E., Newman, R. A., Quinlan, R. A., Goldman, J. E., Perng, M.-D., Inagaki, M. and Snider, N. T. (2019). Site-specific phosphorylation and caspase cleavage of GFAP are new markers of Alexander disease severity. Elife: e47789.
Category
Stem Cell > Pluripotent stem cell > Cell pluripotency
Cell Biology > Cell engineering > Partial reprogramming
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








