- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Radioactive Assay of in vitro Glutamylation Activity of the Legionella pneumophila Effector Protein SidJ
Published: Vol 10, Iss 19, Oct 5, 2020 DOI: 10.21769/BioProtoc.3770 Views: 3803
Reviewed by: David A. CisnerosSrujana Samhita YadavalliCristina Colomer-Winter

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

In vitro and in vivo Assessment of Protein Acetylation Status in Mycobacteria
Krishna K. Singh [...] Deepak K. Saini
Jul 5, 2019 5666 Views

In vitro Glutamylation Inhibition of Ubiquitin Modification and Phosphoribosyl-Ubiquitin Ligation Mediated by Legionella pneumophila Effectors
Alan G. Sulpizio [...] Yuxin Mao
Nov 5, 2020 3920 Views

Detection and Analysis of S-Acylated Proteins via Acyl Resin–Assisted Capture (Acyl-RAC)
Dina A. Abdulrahman and Michael Veit
Apr 5, 2025 1834 Views
Abstract
The Legionella effector protein SidJ has recently been identified to perform polyglutamylation on another Legionella effector, SdeA, ablating SdeA’s activity. SidJ is a kinase-like protein that requires the small eukaryotic protein calmodulin to perform glutamylation. Glutamylation is a relatively uncommon type of post-translational modification, where the amino group of a free glutamate amino acid is covalently linked to the γ-carboxyl group of a glutamate sidechain in a substrate protein. This protocol describes the SidJ glutamylation reaction using radioactive [U-14C] glutamate and its substrate SdeA, the separation of proteins by gel electrophoresis, preparation of gels for radioactive exposure, and relative quantification of glutamylation activity. This procedure is useful for the identification of substrates for glutamylation, characterization of substrate and glutamylase activities due to mutations, and identification of proteins with glutamylation activity. Some studies have assayed glutamylation with the use of [3H] glutamate (Regnard et al., 1998) and the use of the GT335 antibody (Wolff et al., 1992). However, the use of [U-14C] glutamate requires a shorter radioactive exposure time with no dependence on antibody specificity.
Keywords: SidJBackground
Legionella pneumophila is an infectious bacteria that causes Legionnaires’ disease (McDade et al., 1977), a potentially fatal form of pneumonia. During infection, Legionella utilizes an arsenal of over 300 effector proteins, many having unusual, unidentified biochemical functions that act to hijack host cellular functions (Hubber and Roy, 2010). One process co-opted by Legionella is the ubiquitination system. Studies have demonstrated that the SidE family of proteins can perform phosphoribosyl ubiquitination of substrate proteins independent of E1 and E2 enzymes (Bhogaraju et al., 2016; Qiu et al., 2016; Kotewicz et al., 2017). Some studies have implicated the importance of SidJ in the regulation of the SidE family of proteins, but the mechanism of regulation was not identified (Havey and Roy, 2015; Jeong et al., 2015; Urbanus et al., 2016). It was previously suggested that SidJ may act as a phosphoribosyl deubiquitinase (Qiu et al., 2017) using Legionella purified SidJ (Qiu and Luo, 2019); however, recent studies do not replicate these results (Bhogaraju et al., 2019; Wan et al., 2019; Shin et al., 2020). Our group (Sulpizio et al., 2019), and others (Bhogaraju et al., 2019; Black et al., 2019; Gan et al., 2019), have recently demonstrated that SidJ can polyglutamylate the SidE family member SdeA. For verification of this activity, it was important to recapitulate these findings in an in vitro reaction.
SidJ has a C-Terminal IQ helix, that can bind the eukaryotic protein calmodulin in a calcium-independent manner. Using this binding ability, the structure of SidJ in complex with calmodulin was determined by X-ray crystallography. SidJ contains a kinase-like domain with structural homology to many conserved features found in kinases. This kinase domain is positioned in an active conformation through interactions with calmodulin. Based on these features of SidJ, reaction components were identified and used for in vitro glutamylation assays. Other assays have been developed using radioactive glutamate and gel extraction of modified substrates with liquid scintillation to detect modification (Black et al., 2019). Liquid scintillation and mass spectrometry may provide more precise quantification of the amount of modified substrate and for the number of glutamates attached to a substrate side-chain, respectively. The assay described in this protocol allows for visualization of activity by autoradiogram and relative quantitation of activity. This assay can be used to identify SidJ substrates and analyze the effect of point mutations on activity. In addition, this protocol may also be used to identify other proteins or pseudokinases that can function as glutamylases.
Materials and Reagents
- Kim wipes (Kimberly-Clark Professional, catalog number: 34120 )
- Gloves (VWR, catalog number: 89038-270 )
- Film wrap (Spring Grove, catalog number: 405618 )
- Filter paper (GE Healthcare Life Sciences, Whatman GB003, catalog number: 10547922 )
- Laboratory tape (VWR, catalog number: 89098-062 )
- Pipette tips:
10 μl XL Graduated Tips (USA Scientific, Tip One, catalog number: 1110-3700 )
200 μl Graduated Quick Rack (Laboratory Products Sales, catalog number: 130430 )
1,250 μl Pipette tips (Laboratory Product Sales, catalog number: L134770 ) - 1.7 ml Microtubes (Corning Incorporated, Axygen, catalog number: MCT-175-C)
- 50 ml Centrifuge tubes (VWR, catalog number: 525-0637 )
- Recombinant proteins: SidJ 89-853 truncation, SdeA 211-1152 truncation, and human calmodulin 2. Proteins were expressed with an N-terminal 6xHis Sumo tag in Escherichia coli Rosetta cells and purified as described previously (Sulpizio et al., 2019). Final purified proteins were stored in a buffer (20 mM Tris pH 7.5, 150 mM NaCl) without glycerol, aliquoted, flash-frozen, and stored at -80 °C
- Glutamic acid, L-[14C(U)] 50 μCi (Perkin Elmer, catalog number: NEC290E050UC ), stored at -20 °C, the manufacturer suggests 4 °C
- 2-Mercaptoethanol (Sigma, catalog number: M3148-100ML )
- 2-Propanol (J.T.Baker, catalog number: 9079-03 )
- 30% Acrylamide/Bis solution 37.5:1 (Bio-Rad, catalog number: 1610158 )
- Acetic acid, glacial (J.T. Baker, catalog number: 9508-06 )
- Adenosine 5′-triphosphate disodium salt hydrate (Sigma, catalog number: A2382-10G )
- Ammonium persulfate (APS) (Amresco, catalog number: 0486-100G )
- Brilliant Blue R-250 (Fisher, catalog number: BP101-50 )
- Bromophenol Blue sodium salt (Fisher, catalog number: BP114-25 )
- DL-Dithiothreitol (DTT) (Amresco, catalog number: M109-25g )
- Ethanol 200-proof (Koptec, catalog number: V1001 )
- Glycerol (Mallinckrodt Chemicals, catalog number: 5092-16 )
- Glycine (VWR, catalog number: 0167-5KG )
- Magnesium chloride, 6-hydrate (Mallinckrodt Chemicals, catalog number: 5958-04 )
- Methanol (Fisher, catalog number: A454SK-4 )
- N,N,N’,N’-tetramethylethylene-diamine (TEMED) (Bio-Rad, catalog number: 161-0800 )
- Precision Plus Protein All Blue Standards Protein Ladder (Bio-Rad, catalog number: 161-0373 )
- Sodium chloride (VWR, catalog number: 0241-10KG )
- Sodium dodecyl sulfate (SDS) (VWR Life Sciences, catalog number: 0227-1KG )
- Tris (VWR, catalog number: 0497-5KG )
- Reaction Buffer (see Recipes)
- 1 M MgCl2 Solution (see Recipes)
- 100 mM ATP Solution, pH 7.5 (see Recipes)
- 10x SDS-PAGE Running Buffer (see Recipes)
- SDS Sample Buffer (see Recipes)
- Coomassie Stain (see Recipes)
- Coomassie Destaining Solution (see Recipes)
- SDS-PAGE Gel (see Recipes)
12% Resolving Gel
4% Stacking Gel
Note: Products were stored as suggested by manufacture except where listed.
Equipment
- -80 °C freezer (So-Low, model: PV85-21 )
- Exposure cassette (GE Healthcare Life Sciences, model: 63003545 )
- Fixed speed centrifuge (Benchmark, model: myFugeTM mini centrifuge, Type: C1008-C)
- Fluorescent image analyzer (FujiFilm Corporation, GE Healthcare Biosciences, model: Typhoon FLA 7000 )
- Offset Flat-Tip Forceps (Fisher, model: 16-100-116 )
- Gel dryer (Bio-Rad, model: 583 )
- Gel electrophoresis apparatus (Bio-Rad, model: Mini-PROTEAN Tetra System )
- Gel electrophoresis power supply (Bio-Rad, model: PowerPac Basic )
- Gel imager (Bio-Rad, model: Chemidoc MP Imaging System )
- Ice bucket
- Imaging plate (Fuji Film, model: FUJI BAS-IP MS 2025 )
- Labcoat (VWR, catalog number: 10141-306 )
- Lightbox (Laboratory Supplies Company Inc., model: G129A )
- Microwave (Sharp, model: R230KW )
- Pipettes (Gilson Pipetman classic P2, P20, P200, P1000, catalog numbers: F144801, F123600, F123601, F123602)
- Pyrex container (secondary containment)
- Refrigerate benchtop centrifuge (International Equipment Company, model: Micromax RF )
- Rocking shaker (Reliable Scientific, model: 55D 12x16 )
- Scissors
- Sheet protectors (Clear file, Archival Plus 5x7 Print, catalog number: 370100B)
- Vacuum pump (Bio-Rad, model: HydroTech Vacuum Pump )
- Vortex mixer 120V (Corning LSE, model: 6775 )
- Water bath incubator (PolyScience, model: WB05 )
Software
- Fiji Version 1.0 (Citation Schindelin et al., 2012, http://fiji.sc)
- Typhoon FLA 7000 Control Software Version 1.2 (General Electric Company, www.gelifesciences.com/contact/)
- GraphPad Prism 7 for Mac OS X (GraphPad Software, Inc., www.graphpad.com)
Procedure
- SidJ in vitro glutamylation reaction
- Review the flowchart of the experimental outline for SidJ radioactive glutamylation before beginning procedure (Figure 1).
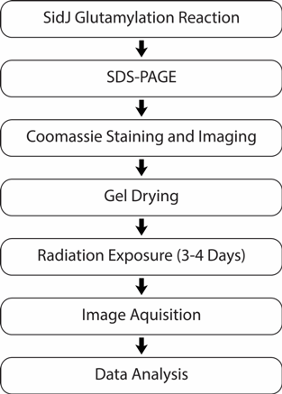
Figure 1. Experimental Outline for SidJ Radioactive Glutamylation. A flowchart of general experimental steps described in this procedure beginning with the SidJ radioactive glutamylation assay and concluding with data analysis. - Thaw recombinantly purified SidJ 89-853, SdeA 211-1152, and calmodulin on ice. Thaw radioactive glutamic acid stock at room temperature in a radioactive workspace.
Note: When working with radioactive materials, ensure that all regulations are followed. Use proper PPE, maximize distance from the sample, and minimize exposure time. - Warm water bath to 37 °C and chill centrifuge to 4 °C (if possible).
- Prepare stock solutions, on ice, listed in Table 1 by diluting in reaction buffer (20 mM Tris pH 7.5, 50 mM NaCl).
Table 1. Stock solution concentrations for SidJ in vitro glutamylation reaction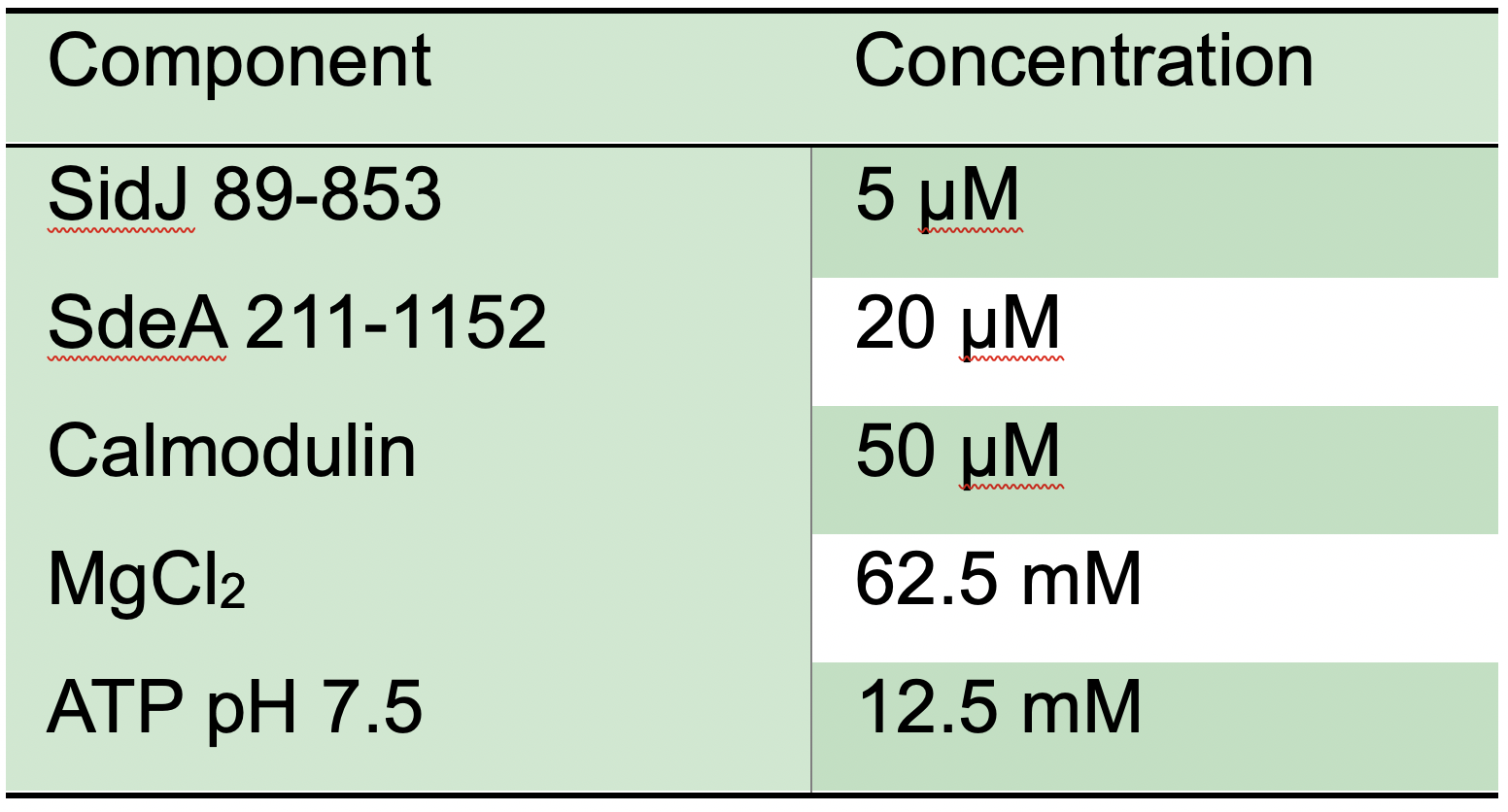
- Pipette the volumes of stock solution for a single 12.5 μl reaction listed in Table 2 into a chilled 1.7 ml microcentrifuge tube on ice. Move to radioactive workspace before the addition of [U-14C] glutamic acid. Pipette ATP last to initiate the reaction. Immediately vortex reaction gently, centrifuge briefly (approximately 10 s max speed), and incubate samples at 37 °C in a water bath for 30 min.
- Optional: Prepare a master mix for Step A5. Combine components contained in all reactions by pipetting stock solutions and 3 μl of reaction buffer per reaction. Prepare approximately 10% more reaction mix than needed for samples. The addition of reaction buffer for the master mix dilutes components to maintain protein stability. If a master mix is prepared, for each reaction, subtract the volumes of reaction components included in the mix and 3 μl of reaction buffer from the amount used in Table 2.
Table 2. SidJ in vitro glutamylation reaction components and concentrations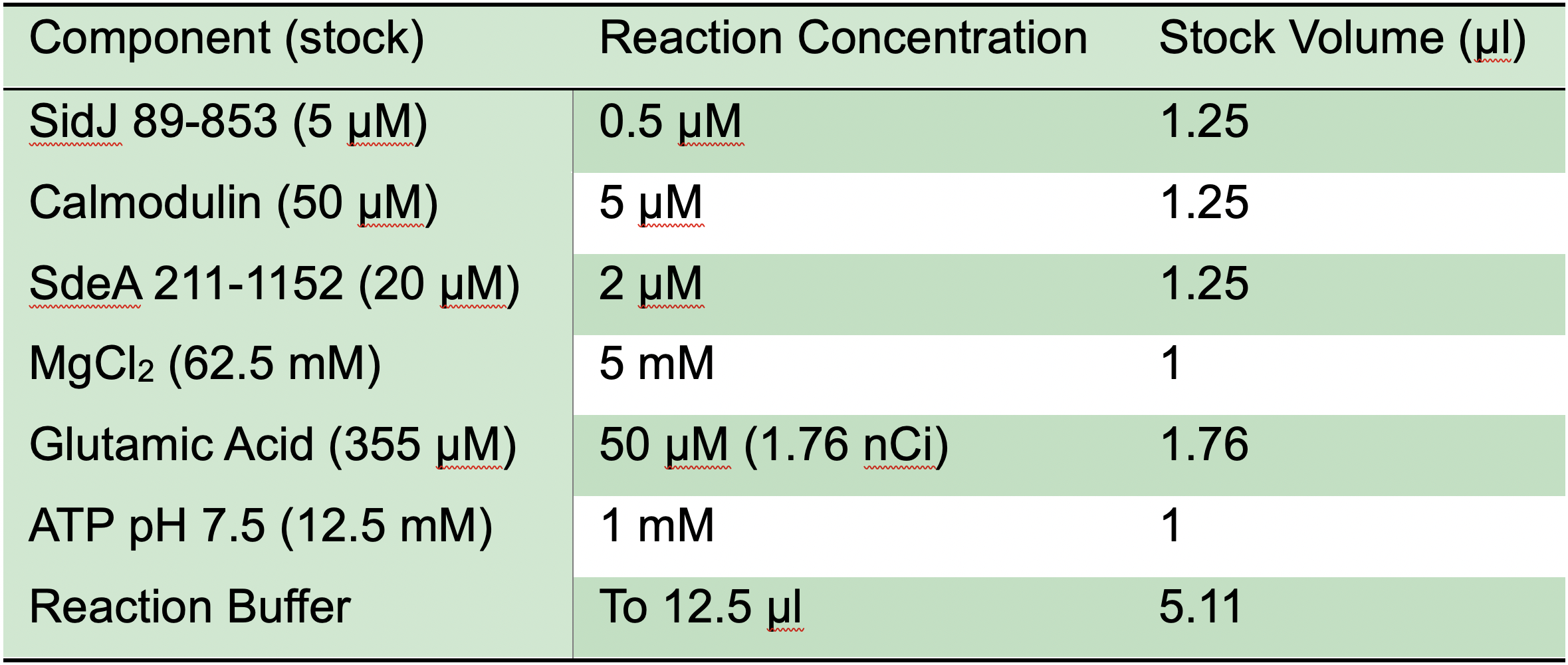
Note: For comparison of samples that may have minor differences in activities, the final protein concentration in the reaction of SidJ can be reduced to 50 nM. If studying the impact of calmodulin-binding on SidJ activity, reducing the final molar concentration of calmodulin to the concentration of SidJ may be beneficial. The reaction time can also be shortened to 15 min. Reducing SidJ concentration may make it difficult to accurately compare protein amounts and therefore relative activity using Coomassie staining. - Stop the reaction with the addition of 3 μl of SDS sample buffer. Vortex to mix and centrifuge briefly (approximately 10 s at max speed).
- Review the flowchart of the experimental outline for SidJ radioactive glutamylation before beginning procedure (Figure 1).
- Gel electrophoresis and preparation for radiation exposure
- Electrophorese 2.5 μl of protein ladder and 13 μl of each reaction using a SDS-PAGE gel (4% separating gel, 12% resolving gel) at 80 V. Once samples have migrated through stacking gel, increase the voltage to 150 V and electrophorese until the dye front reaches the bottom of the gel. The running buffer will likely be contaminated with radioactive material.
- Quickly remove the gel from casting glass and remove the stacking gel. Transfer the gel to a plastic container with a lid in secondary containment. Microwave Coomassie stain, enough to cover gel, in a covered microwave-safe container until boiling. Pour heated Coomassie stain into the plastic container with the gel, ensuring not to inhale fumes, and quickly cover container.
- Stain gel by rocking using rocking shaker for a few hours, to overnight, at room temperature.
Note: There was some difficulty visualizing the calmodulin band using Coomassie staining. Staining temperature and time may be decreased if calmodulin can be adequately stained. - After staining, discard stain into a radioactive liquid waste container. Rinse with the destaining solution and then incubate in destaining solution for approximately 30 min to 1 h. Then discard the solution and repeat incubation. Repeat until protein bands are visible. If some background staining persists, allow longer water destaining in Step B5.
- Rehydrate gel and destain further by incubation in ddH2O while rocking for 1-2 h. Remove water and repeat if necessary. Adding a Kim wipe can assist in destaining and provide cleaner gel images.
- After the gel is rehydrated, transfer the gel to a sheet protector and image using a gel imager. Wiping dust and staining imperfections with a Kim wipe may help obtain clearer images (Figure 2A)
- Cut a sheet of filter paper in half with scissors. Gently transfer gel to two stacked layers of filter paper, by touching one side of the gel to the paper, and then smoothly allowing it to lay flat. Avoid bubbles and imperfections. Cover with a layer of plastic wrap and smooth out any imperfections. If there are lines in the plastic wrap, this may impact exposure efficiency.
- Dry the gel using a vacuum gel dryer at 80 °C for 1 h or until the gel is completely dried. Ensure gel is dried before removing vacuum to prevent gel cracking.
- While drying the gel, erase the image plate by exposing it on the lightbox for 30 min.
- Wrap plastic wrap around the filter paper to prevent damage to the image plate. Tape the gel and two filter paper sheets to the bottom side of the exposure cassette. Avoid using thick plastic sheet protectors as they will reduce signal.
- Place image plate over gel with the white side facing the gel. Record the positioning of the notched edge of the image plate relative to the gel. Expose for 3 to 4 days to obtain optimal signal intensity.
- Image acquisition
- After exposure, minimize the ambient light in the imaging room to prevent the alteration of the image signal. Attach the black magnetic section of the image plate to the phosphor stage and attach the stage to the imager.
- Open the software and select the phosphorimaging option. Select the following parameters for image acquisition: Laser: 650 nm, Filter: [IP], PMT: 1000, Pixel Size: 25 μM, Latitude: L5, Stage: Phosphor Stage, Mode: All.
- Click the “Start Scan” button and allow the instrument to scan the image plate. Once, the area containing your gel has been scanned acquisition can be stopped.
If a manual selection was chosen, ensure that the area containing the gel was accurately selected. Partial scanning of gels will prevent accurate comparison between portions of the gel scanned separately. - A gel file will then be saved in the area chosen upon scanning. This can be saved to a flash drive and transported to a personal computer (Figure 2B).
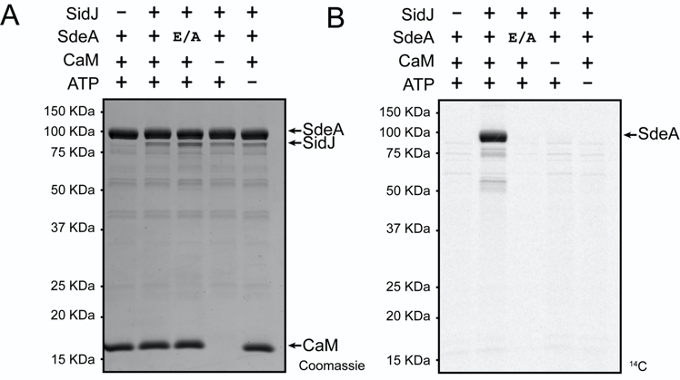
Figure 2. Reaction components required for SidJ mediated glutamylation of SdeA. A. SidJ was incubated with the indicated reaction components with concentrations listed in Table 2. The SdeA E/A is a SdeA E860A mutant of the residue identified to be glutamylated by mass spectrometry. Reactions were conducted for 30 min at 37 °C. Proteins were electrophoresed by SDS-PAGE and stained with Coomassie stain. CaM is an abbreviation for calmodulin. B. An autoradiogram of the gel in A that was exposed for 4 days. This figure is from the original research article (Sulpizio et al., 2019).
Data analysis
- Open Fiji, or ImageJ, software. In the menu bar, select “File -> Open” and browse to the location of the .gel file.
If the gel is not horizontally level, select “Image -> Transform -> Rotate”. Select the “Preview” checkbox and manually input a degree rotation and alter until gel preview is level. Then click “OK”. - In the toolbar, select the rectangle selection tool. In the image window, click and drag to create a rectangular selection. This selection area should be large enough to fit the largest band. Then move this selection to cover the first band you would like to measure. In the menu bar, click “Analyze -> Measure.” A Results pop up window will appear with intensity data. Click in the center of this rectangle and drag it to surround the next band to be analyzed and measure intensity. Repeat process, recording which measurements correspond to which samples until the intensity of all bands has been measured. Measure the intensity of a background selection, ideally on the area where the gel was exposed but no samples were electrophoresed. All rectangular selections should be identical for an accurate comparison of intensities. Click on the Results window and in the menu bar select “File -> Save As…” and choose a file name and location.
- Measurements of relative radiographic intensities of bands can then be calculated by multiplying the area of selection by the mean, to get total intensity, for all measurements. Subtract the background measurement from all samples. Relative intensities can be compared by dividing one sample’s total intensity by another sample’s total intensity. Calculation of relative intensities should only be made for samples exposed on the same image plate, and preferably electrophoresed within the same gel. For example, the intensity of protein mutants can be divided by the wild type total radiographic intensity to ascertain the effect of these mutants on activity. Removal or alteration of reaction components can also be compared.
- Relative radiographic intensities can then be displayed using a bar graph. Assays and quantification should be repeated in triplicate. Error bars can be represented as a standard deviation, and P-values were calculated with a single-tailed t-test. For examples of data used for graphing and statistical calculations see “Additional Files, Source Data 1” (Sulpizio et al., 2019).
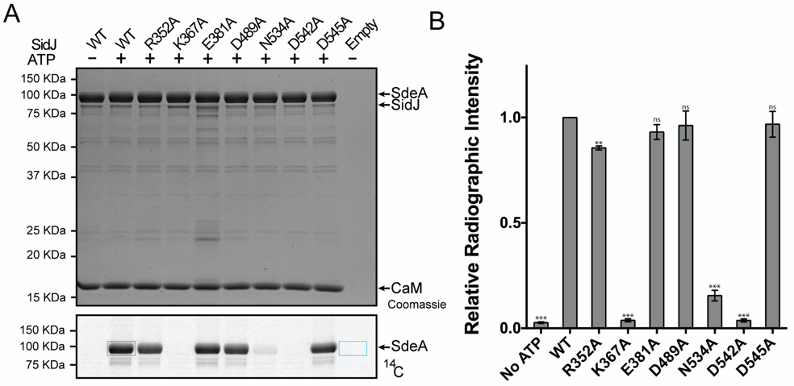
Figure 3. Quantification of SidJ kinase-like domain mutants. A. SidJ and SidJ mutants were incubated with wild type SdeA and reaction components, with concentrations listed in Table 2, in the presence or absence of ATP for 15 min at 37 °C. The lane labeled Empty contained SDS-Sample Buffer with reaction buffer substituted for sample. Top panel: Coomassie stained gel, Bottom Panel: Autoradiogram with an example of a region used for WT signal quantification and the region for background subtraction shown as red and cyan boxes, respectively. WT and CaM are abbreviations for wild type and calmodulin, respectively. B. Quantitation of relative radioactive intensity of reactions of the bottom panel of A. Bar graphs are averages of three separate experiments with error bars depicted as standard deviation. The P-values were calculated from a t-test and ns, not significant, **, P < 0.01, ***, P < 0.001. The graph was generated using GraphPad Software. This figure is from original research article (Sulpizio et al., 2019).
Recipes
- Reaction Buffer
50 mM Tris pH 7.5
50 mM NaCl
Stored at room temperature - 1 M MgCl2 Solution
Stored at room temperature - 100 mM ATP Solution, pH 7.5
Adjust pH to 7.5, aliquot and store at -80 °C - 10x SDS-PAGE Running Buffer
Store at room temperature and dilute 10-fold with ddH2O for useComponent 1 L Tris-Base 30 g Glycine 140 g SDS 10 g ddH2O To 1 L - SDS Sample Buffer
Store at room temperature, freeze aliquots -20 °C for extended storageComponent Concentration Bromophenol Blue 0.25%(w/v) DTT 0.5 M Glycerol 50% (v/v) SDS 10% (w/v) - Coomassie Stain
Store at room temperatureComponent 500 ml Methanol 225 ml ddH2O 225 ml Glacial Acetic Acid 50 ml Brilliant Blue R250 1.25 g - Coomassie Destaining Solution
Store at room temperatureComponent 1 L Ethanol 450 ml ddH2O 450 ml Glacial Acetic Acid 100 ml - SDS-PAGE Gel
- 12% Resolving Gel
Component 8 gels Final Conc. ddH2O 13.3 ml 30% Acrylamide/Bis Solution 16 ml 12% 1.5 M Tris pH 8.8 10 ml 375 mM 10% SDS 400 μl 0.1% 10% APS 300 μl 0.075% TEMED 24 μl 0.06% - 4% Stacking Gel
Component 8 gels Final Conc. ddH2O 11.2 ml 30% Acrylamide/Bis Solution 2.16 ml 4.14% 1.0 M Tris pH 6.8 2 ml 128 mM 10% SDS 160 μl 0.1% 10% APS 110 μl 0.07% TEMED 16 μl 0.1%
- 12% Resolving Gel
Acknowledgments
This work was supported by National Institute of Health (NIH) grant 5R01GM116964 (to YM), the Cornell University Harry and Samuel Mann Outstanding Graduate Student Award (to AGS) and by the NIH under Ruth L Kirschstein National Research Service Award (6T32GM008267) from the NIGMS (to MEM).
Protocol from original research article: Sulpizio, A., M. E. Minelli, M. Wan, P. D. Burrowes, X. Wu, E. J. Sanford, J.-H. Shin, B. C. Williams, M. L. Goldberg, M. B. Smolka and Y. Mao (2019). "Protein polyglutamylation catalyzed by the bacterial calmodulin-dependent pseudokinase SidJ." eLife 8: e51162.
Competing interests
The authors declare no competing interests.
References
- Bhogaraju, S., Bonn, F., Mukherjee, R., Adams, M., Pfleiderer, M. M., Galej, W. P., Matkovic, V., Lopez-Mosqueda, J., Kalayil, S., Shin, D. and Dikic, I. (2019). Inhibition of bacterial ubiquitin ligases by SidJ-calmodulin catalysed glutamylation. Nature 572(7769): 382-386.
- Bhogaraju, S., Kalayil, S., Liu, Y., Bonn, F., Colby, T., Matic, I. and Dikic, I. (2016). Phosphoribosylation of ubiquitin promotes serine ubiquitination and impairs conventional ubiquitination. Cell 167(6): 1636-1649. e1613.
- Black, M. H., Osinski, A., Gradowski, M., Servage, K. A., Pawłowski, K., Tomchick, D. R. and Tagliabracci, V. S. (2019). Bacterial pseudokinase catalyzes protein polyglutamylation to inhibit the SidE-family ubiquitin ligases. Science 364(6442): 787-792.
- Gan, N., Zhen, X., Liu, Y., Xu, X., He, C., Qiu, J., Liu, Y., Fujimoto, G. M., Nakayasu, E. S., Zhou, B., Zhao, L., Puvar, K., Das, C., Ouyang, S. and Luo, Z.-Q. (2019). Regulation of phosphoribosyl ubiquitination by a calmodulin-dependent glutamylase. Nature 572(7769): 387-391.
- Havey, J. C. and Roy, C. R. (2015). 583 " target="_blank">Toxicity and SidJ-mediated suppression of toxicity require distinct regions in the SidE family of Legionella pneumophila effectors. Infect Immun 83(9): 3506-3514.
- Hubber, A. and Roy, C. R. (2010). Modulation of host cell function by Legionella pneumophila type IV effectors. Annu Rev Cell Dev Biol 26: 261-283.
- Jeong, K. C., Sexton, J. A. and Vogel, J. P. (2015). Spatiotemporal regulation of a Legionella pneumophila T4SS substrate by the metaeffector SidJ. PLoS Pathog 11(3): e1004695.
- Kotewicz, K. M., Ramabhadran, V., Sjoblom, N., Vogel, J. P., Haenssler, E., Zhang, M., Behringer, J., Scheck, R. A. and Isberg, R. R. (2017). A single Legionella effector catalyzes a multistep ubiquitination pathway to rearrange tubular endoplasmic reticulum for replication. Cell Host Microbe 21(2): 169-181.
- McDade, J. E., Shepard, C. C., Fraser, D. W., Tsai, T. R., Redus, M. A. and Dowdle, W. R. (1977). Legionnaires' disease: isolation of a bacterium and demonstration of its role in other respiratory disease. N Engl J Med 297(22): 1197-1203.
- Qiu, J. and Luo, Z. Q. (2019). Methods for Noncanonical Ubiquitination and Deubiquitination Catalyzed by Legionella pneumophila Effector Proteins. Methods Mol Biol 1921: 267-276.
- Qiu, J., Sheedlo, M. J., Yu, K., Tan, Y., Nakayasu, E. S., Das, C., Liu, X. and Luo, Z. Q. (2016). Ubiquitination independent of E1 and E2 enzymes by bacterial effectors. Nature 533(7601): 120-124.
- Qiu, J., Yu, K., Fei, X., Liu, Y., Nakayasu, E. S., Piehowski, P. D., Shaw, J. B., Puvar, K., Das, C., Liu, X. and Luo, Z.-Q. (2017). A unique deubiquitinase that deconjugates phosphoribosyl-linked protein ubiquitination. Cell Res 27(7): 865-881.
- Regnard, C., Audebert, S., Desbruyères, É., Denoulet, P. and Eddé, B. (1998). Tubulin polyglutamylase: partial purification and enzymatic properties. Biochemistry 37(23): 8395-8404.
- Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., Preibisch, S., Rueden, C., Saalfeld, S., Schmid, B., Tinevez, J.-Y., White, D. J., Hartenstein, V., Eliceiri, K., Tomancak, P. and Cardona, A. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods 9(7): 676-682.
- Shin, D., Mukherjee, R., Liu, Y., Gonzalez, A., Bonn, F., Liu, Y., Rogov, V. V., Heinz, M., Stolz, A., Hummer, G., Dötsch, V., Luo, Z.-Q., Bhogaraju, S. and Dikic, I. (2020). Regulation of phosphoribosyl-linked serine ubiquitination by deubiquitinases DupA and DupB. Mol Cell 77(1): 164-179. e166.
- Sulpizio, A., Minelli, M. E., Wan, M., Burrowes, P. D., Wu, X., Sanford, E. J., Shin, J. H., Williams, B. C., Goldberg, M. L., Smolka, M. B. and Mao, Y. (2019). Protein polyglutamylation catalyzed by the bacterial calmodulin-dependent pseudokinase SidJ. eLife 8: e51162.
- Urbanus, M. L., Quaile, A. T., Stogios, P. J., Morar, M., Rao, C., Di Leo, R., Evdokimova, E., Lam, M., Oatway, C., Cuff, M. E., Osipiuk, J., Michalska, K., Nocek, B. P., Taipale, M., Savchenko, A. and Ensminger, A. W. (2016). Diverse mechanisms of metaeffector activity in an intracellular bacterial pathogen, Legionella pneumophila. Mol Syst Biol 12(12): 893.
- Wan, M., Sulpizio, A. G., Akturk, A., Beck, W. H. J., Lanz, M., Faca, V. M., Smolka, M. B., Vogel, J. P. and Mao, Y. (2019). Deubiquitination of phosphoribosyl-ubiquitin conjugates by phosphodiesterase-domain-containing Legionella effectors. Proc Natl Acad Sci U S A 116(47): 23518-23526.
- Wolff, A., de Nechaud, B., Chillet, D., Mazarguil, H., Desbruyeres, E., Audebert, S., Edde, B., Gros, F. and Denoulet, P. (1992). Distribution of glutamylated alpha and beta-tubulin in mouse tissues using a specific monoclonal antibody, GT335. Eur J Cell Biol 59(2): 425-432.
Article Information
Copyright
Sulpizio et al. This article is distributed under the terms of the Creative Commons Attribution License (CC BY 4.0).
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Sulpizio, A. G., Shin, J., Minelli, M. E. and Mao, Y. (2020). Radioactive Assay of in vitro Glutamylation Activity of the Legionella pneumophila Effector Protein SidJ. Bio-protocol 10(19): e3770. DOI: 10.21769/BioProtoc.3770.
- Sulpizio, A., Minelli, M. E., Wan, M., Burrowes, P. D., Wu, X., Sanford, E. J., Shin, J. H., Williams, B. C., Goldberg, M. L., Smolka, M. B. and Mao, Y. (2019). Protein polyglutamylation catalyzed by the bacterial calmodulin-dependent pseudokinase SidJ. eLife 8: e51162.
Category
Microbiology > Microbial biochemistry > Protein > Modification
Microbiology > Microbial biochemistry > Protein > Labeling
Biochemistry > Protein > Posttranslational modification
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









