- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
RNA Stability Measurements Using RT-qPCR in Arabidopsis Seedlings
Published: Vol 10, Iss 14, Jul 20, 2020 DOI: 10.21769/BioProtoc.3680 Views: 6996
Reviewed by: Zhibing LaiAntony ChettoorJason Liang Pin Ng

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

3’ Rapid Amplification of cDNA Ends (3’ RACE) Using Arabidopsis Samples
Encarnación Rodríguez-Cazorla [...] Antonio Vera
Oct 5, 2015 21342 Views

Real-time PCR Analysis of PAMP-induced Marker Gene Expression in Nicotiana benthamiana
Fan Liu [...] Yuanchao Wang
Oct 5, 2018 9304 Views

Laser-Assisted Microdissection and High-Throughput RNA Sequencing of the Arabidopsis Gynoecium Medial and Lateral Domains
Valentín Luna-García and Stefan de Folter
Sep 5, 2024 2066 Views
Abstract
Steady-state mRNA levels are determined by both the rates of transcription and degradation. Regulation of mRNA stability and/or degradation are key factors that can significantly affect mRNA levels and its biological functions. mRNA stability can be measured indirectly after transcription inhibition. This protocol described a rapid and sensitive method of mRNA stability measurement through quantitative reverse transcription PCR (RT-qPCR) after inhibition of RNA transcription by cordycepin in Arabidopsis seedlings.
Keywords: RNA StabilityBackground
The regulation of mRNA stability is a key control point in gene expression regulation. mRNA stability has a profound impact on gene expression, molecular and cellular phenotype, and ultimately, plant development, defense and other biological processes. Multiple methods, e.g., Northern blot analysis, in situ hybridization, can be used to measure mRNA stability after transcription inhibition. In this protocol, we describe a rapid and sensitive method to measure mRNA stability through RT-qPCR after inhibition of transcription by cordycepin. Cordycepin or 3'-deoxyadenosine is an adenosine analogue (see Reference 2). 3'-deoxyadenosine can be incorporated into RNA and inhibits transcription elongation and RNA synthesis due to the absence of a hydroxyl moiety at the 3' position (see Reference 6). We have successfully used this convenient and sensitive method to measure the stability of several low-abundant mRNAs, including primary microRNA transcripts, in Arabidopsis seedlings (Jia et al., 2017). Here we present this protocol with detailed experimental procedures and data analysis methods.
Materials and Reagents
- Supplies
- Petri dishes, polystyrene, size 150 mm × 15 mm, sterile
- 6-well clear multiwell plates, sterile
- KimwipesTM
- Low retention tips
- Materials
- Arabidopsis seeds, including control and treatment or mutant groups
It is strongly recommended to include a mutant with defects in mRNA stability as a positive control, e.g., ect2-1(SALK_002225).
- Arabidopsis seeds, including control and treatment or mutant groups
- Reagents
- Liquid nitrogen
- RNaseZapTM (ThermoFisher, catalog number: AM9780 ) or ELIMINase® (DeconLabs, catalog number: 1101 )
- Murashige and Skoog Basal Medium (Sigma-Aldrich, catalog number: M5519 )
- Agar (Sigma-Aldrich, catalog number: A1296 )
- Household bleach (5.25% w/v sodium hypochlorite)
- Tween® 20 (Sigma-Aldrich, catalog number: P9416 )
- Nuclease-free water
- Cordycepin (Sigma-Aldrich, catalog number: C3394 )
- RNA extraction kit (RNeasy Plant Mini Kit) (QIAGEN, catalog number: 74904 )
- cDNA synthesis kit (iScript cDNA Synthesis Kit) (Bio-Rad, catalog number: 1708890 )
- qPCR kit (SYBRTM Green PCR Master Mix) (ThermoFisher, catalog number: 4309155 )
- qPCR primers (Design qPCR primers on Primer3Plus (Untergasser et al., 2012), and order qPCR primers from Integrated DNA Technology)
- Liquid nitrogen
Equipment
- Multichannel pipettes
- Mortar and pestle
- Autoclave
- -20 °C freezer
- Arabidopsis growth room or chamber
- VWR® Standard Orbital Shaker, Model 3500 (VWR, catalog number: 89032-092 )
- Eppendorf Benchtop Centrifuge 5418, non-refrigerated, with Rotor FA-45-18-11 and rotor lid, 230 V/50-60 Hz (Eppendorf, catalog number: 5418000017 )
- Applied BiosystemsTM QuantStudioTM 5 Real-Time PCR System (ThermoFisher, catalog number: A34322 ), or Bio-Rad CFX96 Touch Real-Time PCR Detection System (Bio-Rad, catalog number: 1855196 )
Software
- Primer3Plus (A. Untergasser, https://primer3plus.com/cgi-bin/dev/primer3plus.cgi)
- Excel (Microsoft)
- Applied BiosystemsTM Analysis Software or Thermo Fisher Cloud (Thermo Fisher Scientific, available here)
Procedure
- Prepare reagents and buffers
Note: Calculate the required amount of MS medium and cordycepin before starting reagent preparation based on experimental settings. Approximately 50 ml MS medium is required to prepare one 150 mm plate. Each well of the six-well plate requires around 3 ml MS medium. At least two replicates should be included for each sample at each time point. Depending on the size of the seedlings, approximately 10 to 20 seedlings could fit into one well of the six-well plate.- Prepare 1× MS agar medium plates using 150 mm Petri dishes as described in the ABRC Arabidopsis protocols (Rivero et al., 2014). Prepared plates can be wrapped up and stored at 4 °C for at least a month.
- Prepare 0.5× MS liquid growth medium by making a two-fold dilution of 1× MS medium. Do not add agar. Autoclaved medium can be stored at 4 °C for at least a month.
- Prepare seeds surface-sterilization solution: Use 50% household bleach with distilled water to make solutions with final concentration of 2.6 % w/v sodium hypochlorite, add Tween® 20 to a final concentration of 0.05%.
- Dissolve cordycepin in nuclease-free water to make 0.6 M stock concentration. Store 0.6 M cordycepin stock solution at -20 °C.
- Dilute qPCR primers to 100 μM in nuclease-free water. Store at -20 °C.
- Prepare plant materials
- Sterilize seeds in microcentrifuge tubes by soaking them in the surface-sterilization solution for 5 to 10 min. Remove all bleach by rinsing five to seven times with sterile distilled water (Rivero et al., 2014).
- The Arabidopsis seeds (control and mutant groups, as shown in Figure 1) are sown onto the 1× MS agar plates at a density of about one seed per 1 cm2, facilitating manipulation during subsequent seedling transfer. After cold stratification, the plates are placed vertically in the growth chamber, thus preventing growth of the root into the solid media. Grow Arabidopsis in sterile condition for approximately 12 days following the ABRC Arabidopsis protocols (Rivero et al., 2014).
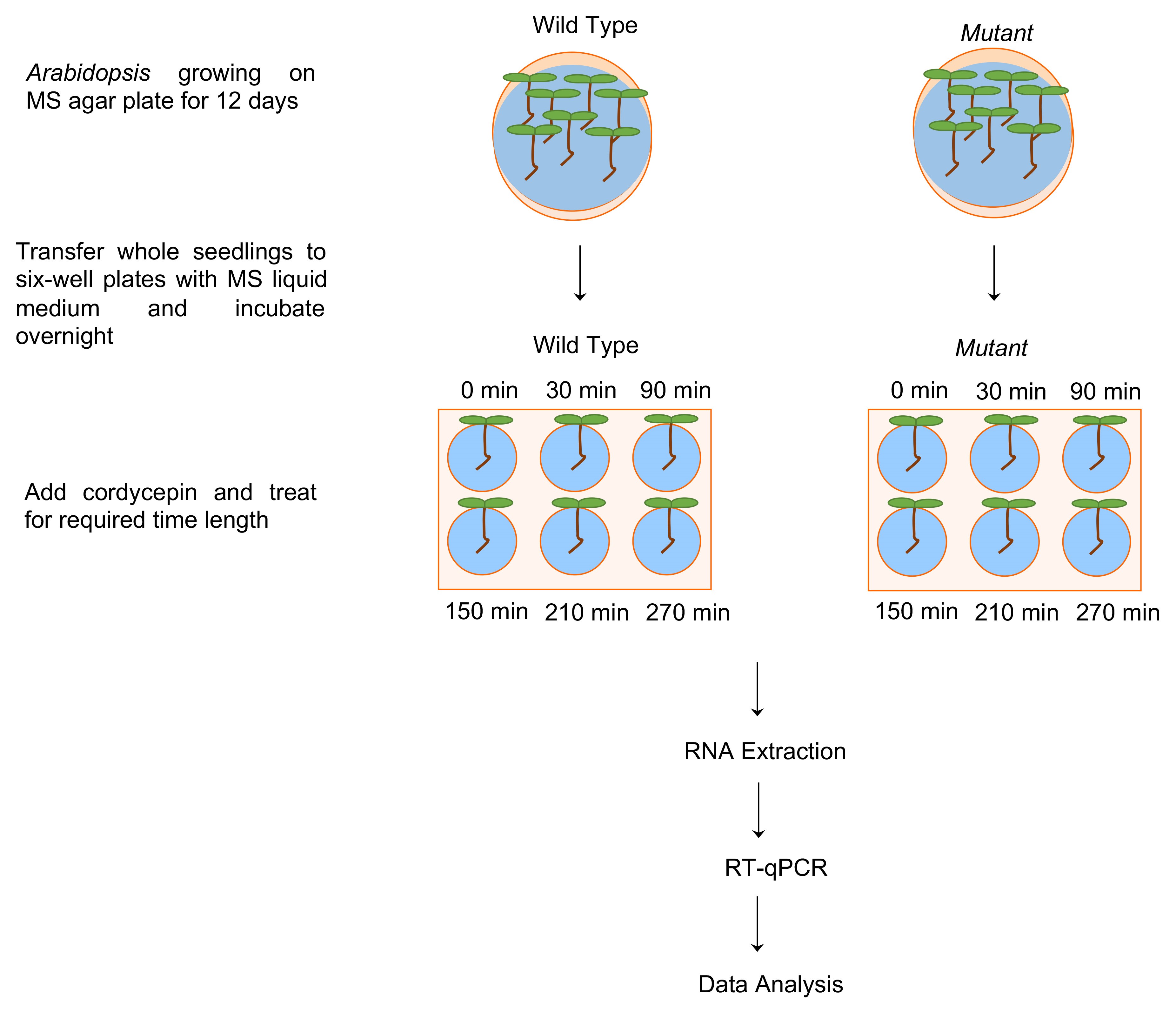
Figure 1. Work flow illustration of RNA stability measurements using RT-qPCR in Arabidopsis. Twelve-day-old Arabidopsis wild type and mutant whole seedlings were transferred from MS medium agar plates to six-well plates with 0.5× MS liquid medium and incubated overnight (Note: Only roots will be submerged in liquid MS). The next day, cordycepin was added to a final concentration of 0.6 mM and the seedlings were collected for RNA extraction at indicated time points after cordycepin treatment. RT-qPCR was then performed to determine RNA levels, and statistical analysis was used to determine the significant difference between wild type and mutant RNA stability.
- Sterilize seeds in microcentrifuge tubes by soaking them in the surface-sterilization solution for 5 to 10 min. Remove all bleach by rinsing five to seven times with sterile distilled water (Rivero et al., 2014).
- Experimental procedures
Day 1- Use sterilized forceps to transfer Arabidopsis whole seedlings to six-well plates with 3 ml 0.5× MS medium per well in tissue culture hood. Be careful not to damage the seedling roots during the transfer.
- Incubate the plates on a shaker at 120 rpm in an Arabidopsis growth room or chamber to equilibrate the seedlings overnight.
Note: Read the RNA extraction kit, cDNA synthesis kit and qPCR kit complete protocols before starting the experiment if using those kits for the first time. Use RNaseZapTM (Invitrogen) or ELIMINase® (DeconLabs) to wipe and decontaminate all pipettes and workstations.- Add cordycepin (Sigma-Aldrich) to a final concentration of 0.6 mM (1 to 1,000 dilution of 0.6 M stock), and treat for required lengths of time (e.g., 0 min, 30 min, 90 min, 150 min, 210 min, 270 min, as shown in Figure 1).
Note: The longest treatment (e.g., 270 min in this experiment) can be performed first, and then add cordycepin based on required lengths of treatment time, and finally collect all samples at the same time (Figures 1 and 2).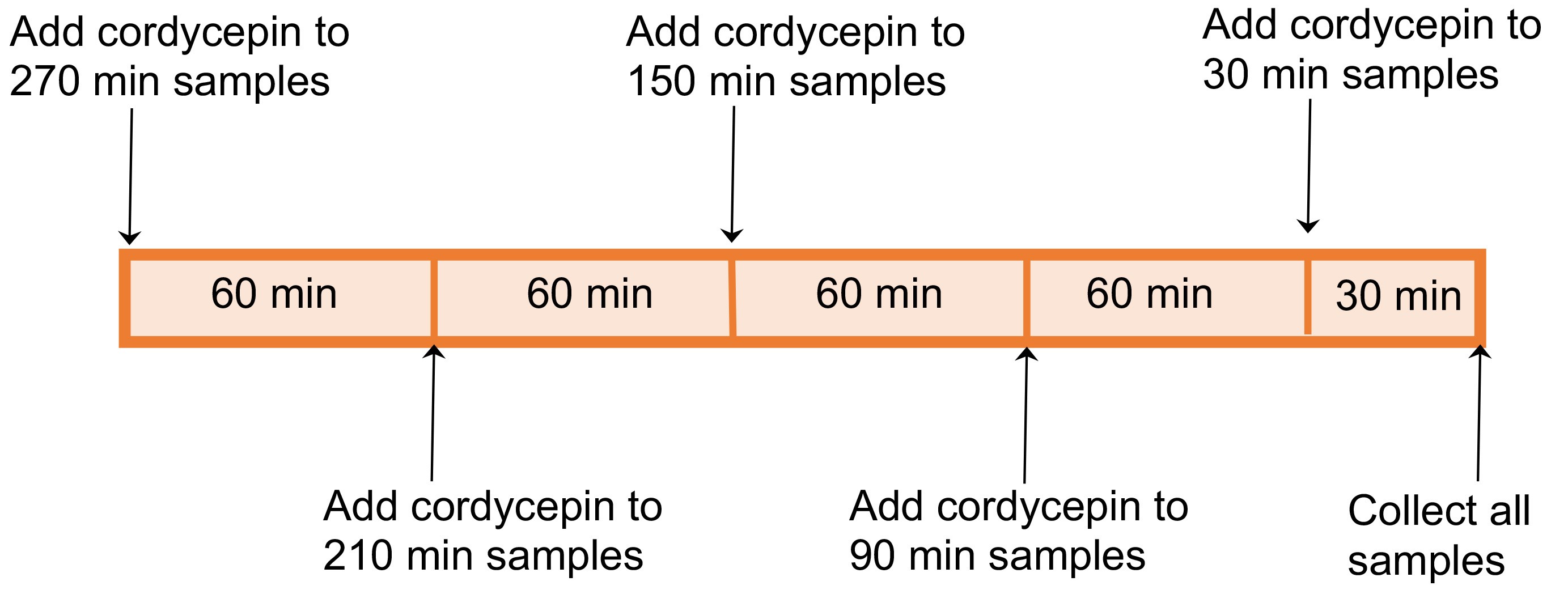
Figure 2. Schematic representation of cordycepin treatment time course analysis - Collect seedlings and blot on KimwipesTM to remove excessive liquid.
- Immediately, freeze the seedlings in liquid nitrogen and grind to a fine powder. Transfer to a cooled, appropriately sized, tube and allow liquid nitrogen to evaporate without allowing the sample to thaw, and then add lysis buffer. Repeat for the remaining seedling samples. Proceed to the next step of RNA extraction as soon as possible following the RNA extraction kit protocol (see Reference 8).
- Perform RNA quality control: Use a Nanodrop or Qubit to determine the RNA sample purity and concentration.
- Perform cDNA synthesis following Bio-Rad iScriptTM cDNA Synthesis Kit protocol (see Reference 3), and qPCR following SYBR® Green PCR Master Mix protocol (see Reference 10).
Note: Negative controls for reverse transcription are required to make sure all DNA are eliminated during RNA extraction. To set up negative controls, replace reverse transcriptase with nuclease-free water during the cDNA synthesis reactions. Include the negative control samples in the following qPCR analysis. qPCR reaction conditions will need to be optimized for different primers, targets or cDNA samples. The amount of the cDNA reaction that is recommended for downstream qPCR is one-tenth of the reaction volume, typically 1 μl for 10 μl reaction. Try to keep Ct values under 35, which indicates strong positive signals. Include at least two housekeeping controls for qPCR analysis. Because qPCR assay is highly sensitive to pipetting errors and variabilities, use multichannel pipettes and low-retention tips to setup qPCR reactions.
- Use sterilized forceps to transfer Arabidopsis whole seedlings to six-well plates with 3 ml 0.5× MS medium per well in tissue culture hood. Be careful not to damage the seedling roots during the transfer.
Data analysis
- Quality control: Make sure the amplification curve is typical, melting curve shows a single peak indicating a single amplification product from each pair of primers (if melting curve analysis was not performed, run an agarose gel to check the amplification product for single amplification), coefficient of variation in replicate group is equal to or less than 10%, and no signal was detected in the negative control groups (Figure 3). If using QuantStudioTM, review the quality flags using Applied BiosystemsTM Analysis Software or Thermo Fisher Cloud (see Reference 1). Remove poor quality or flagged values.
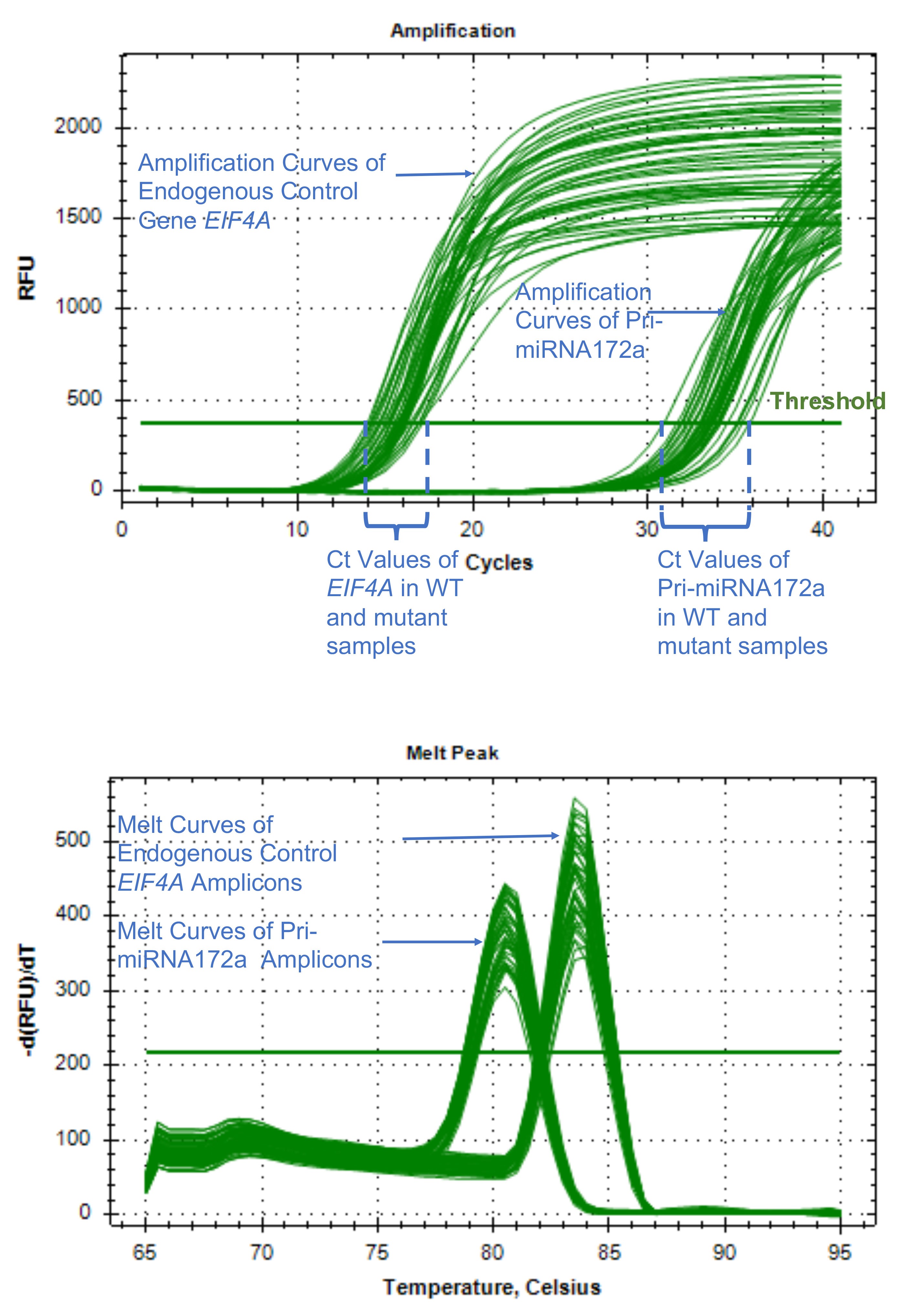
Figure 3. Typical amplification curves and melting curves for an endogenous control and a pri-miRNA gene. Normal amplification curves are smooth during the exponential phase of the PCR. The threshold cycle (Ct) value is the cycle at which the fluorescent signal of the reaction crosses the threshold, which is usually automatically set by the qPCR software at where signal significantly increases over the background. A good melting curve shows a single peak indicating a single amplification product from each pair of primers. The clustering of the curves should be tight for replicates. - Use comparative Ct (△△CT) method to determine relative quantity of each targets in test samples via either software analysis or manual calculation in excel (see Reference 1, Schmittgen and Livak, 2008). The targets are normalized to the endogenous control (e.g., GAPDH, ACTIN, UBQ etc.), and the control or reference sample. The relative quantification or fold change of each target is plotted against time points (Figure 4, Schmittgen and Livak, 2008).
- Statistical analysis: Student’s t-test was performed to check the difference between control and mutant groups at each time point. The cutoff is set as P value < 0.05 and fold change more than 2.
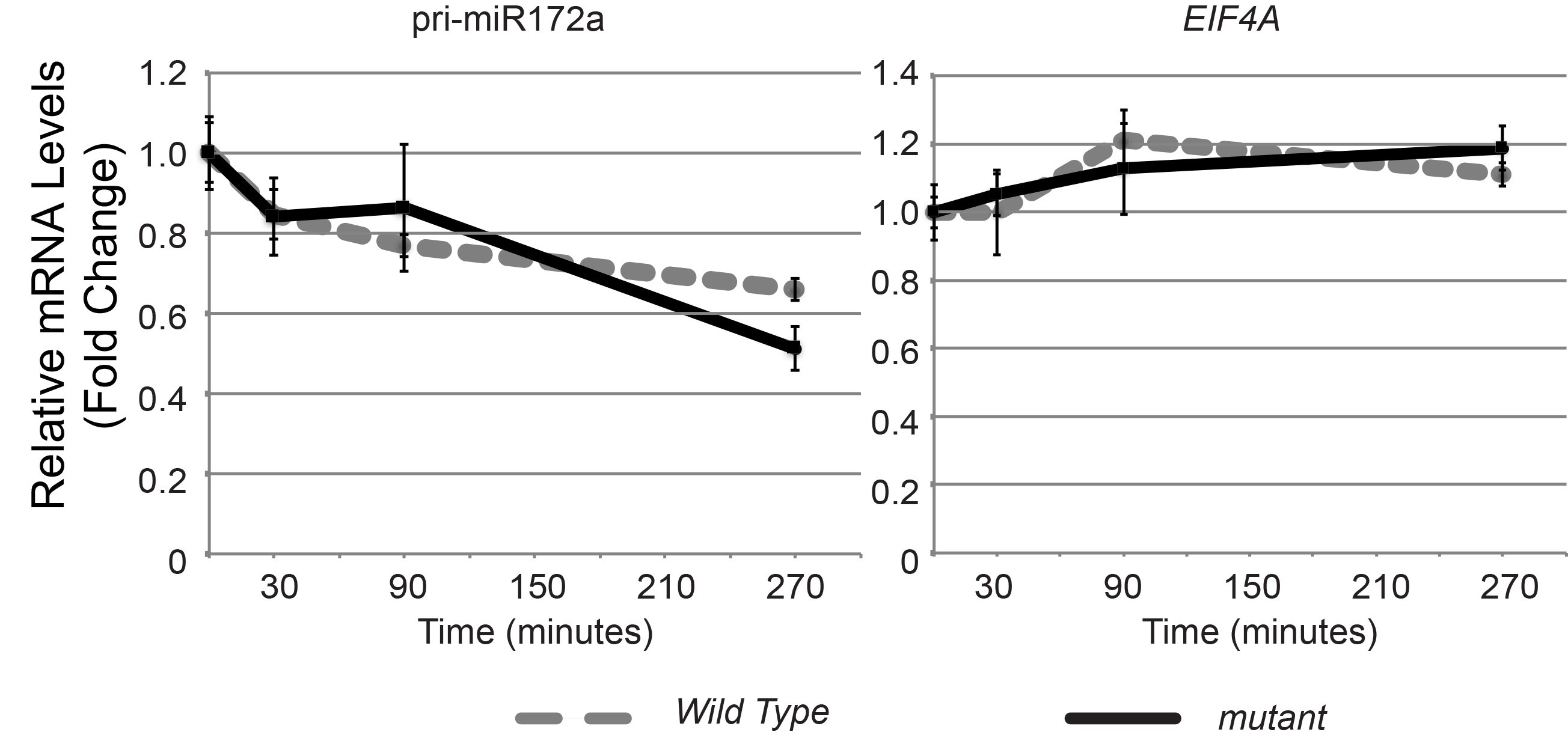
Figure 4. Representative results of RNA stability measurements for pri-miR172a, and EIF4A mRNA. UBQ5 served as an internal control. Samples collected at time point 0 served as the sample control or reference here. Values at time 0 were set to 1. Error bars indicate standard deviation from two sample replicates. For each sample replicate, technical triplicates were performed for individual gene target.
Acknowledgments
We thank Dr. Xuemei Chen and UC Riverside for supporting this work. This work was funded by the National Institutes of Health (GM061146) and Gordon and Betty Moore Foundation (GBMF3046) to Dr. Xuemei Chen. This protocol was adapted from previous works by Lidder et al. (2005), Zhang et al. (2014), and Jia et al. (2017).
Competing interests
The authors declare that there is no conflicts of interest or competing interests.
References
- Applied BiosystemsTM Analysis Software: Applied BiosystemsTM Relative Quantitation Analysis Module USER GUIDE. Thermo Fisher Scientific. Retrieved from: https://assets.thermofisher.com/TFS-Assets/LSG/manuals/MAN0014820_RelativeQuant_UG.pdf.
- Cordycepin: Product Information. Sigma-Aldrich. Retrieved from: https://www.sigmaaldrich.com/content/dam/sigma-aldrich/docs/Sigma/Product_Information_Sheet/c3394pis.pdf.
- iScriptTM cDNA Synthesis Kit: iScriptTM cDNA Synthesis Kit Product Insert, Ver D. Bio-Rad. Retrieved from: https://www.bio-rad.com/webroot/web/pdf/lsr/literature/4106228.pdf.
- Jia, T., Zhang, B., You, C., Zhang, Y., Zeng, L., Li, S., Johnson, K. C. M., Yu, B., Li, X. and Chen, X. (2017). The Arabidopsis MOS4-associated complex promotes microRNA biogenesis and precursor messenger RNA splicing. Plant Cell 29: 2626-2643.
- Lidder, P., Gutiérrez, R. A., Salomé, P. A., McClung, C. R. and Green, P. J. (2005). Circadian control of messenger RNA stability. Association with a sequence-specific messenger rna decay pathway. Plant Physiol 138 (4): 2374-2385.
- NCI Drug Dictionary. Retrieved from: https://www.cancer.gov/publications/dictionaries/cancer-drug/def/cordycepin.
- Rivero, L., Scholl, R., Holomuzki, N., Crist, D., Grotewold, E. and Brkljacic, J. (2014). Handling Arabidopsis plants: growth, preservation of seeds, transformation, and genetic crosses. Methods Mol Biol 1062: 3-25.
- RNeasy Plant Mini Kit: RNeasy Mini Handbook - (EN). QIAGEN. Retrieved from: http://www.bea.ki.se/documents/EN-RNeasy%20handbook.pdf.
- Schmittgen, T. D. and Livak, K. J. (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3(6): 1101 -1108.
- SYBR® Green PCR Master Mix: User Guide SYBR® Green PCR Master Mix and SYBR Green RT-PCR® Reagents Kit. Life Biotechnologies. Retrieved from: https://assets.thermofisher.com/TFS-Assets/LSG/manuals/cms_041053.pdf.
- Untergasser, A., Cutcutache, I., Koressaar, T., Ye, J., Faircloth, B. C., Remm, M. and Rozen, S. G. (2012). Primer3--new capabilities and interfaces. Nucleic Acids Res 40(15): e115.
- Zhang, S., Liu, Y. and Yu, B. (2014). PRL1, an RNA-Binding Protein, Positively Regulates the Accumulation of MiRNAs and SiRNAs in Arabidopsis. PLoS Genet 10 (12): e1004841.
Article Information
Copyright
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Jia, T. and Le, B. H. (2020). RNA Stability Measurements Using RT-qPCR in Arabidopsis Seedlings. Bio-protocol 10(14): e3680. DOI: 10.21769/BioProtoc.3680.
Category
Plant Science > Plant molecular biology > RNA > Transcription
Biochemistry > RNA > RNA structure
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








