- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Pancreatic Acinar Cell Preparation for Oxygen Consumption and Lactate Production Analysis
Published: Vol 10, Iss 10, May 20, 2020 DOI: 10.21769/BioProtoc.3627 Views: 4994
Reviewed by: Khyati Hitesh ShahVanesa Olivares-IllanaWoojong Lee

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Generation of Human iPSC-derived Neural Progenitor Cells (NPCs) as Drug Discovery Model for Neurological and Mitochondrial Disorders
Annika Zink [...] Alessandro Prigione
Mar 5, 2021 9314 Views

Optimization of Extracellular Flux Assay to Measure Respiration of Anchorage-independent Tumor Cell Spheroids
Zaineb Javed [...] Nadine Hempel
Feb 20, 2022 4576 Views

Mito Stress Assay of PBMCs With Seahorse XFe96 Flux Analyzer and Comparison of Poly-D-Lysine and Poly-L-Lysine for Cell Affinity
Kumudu Subasinghe [...] Nicole Phillips
Jun 5, 2025 1975 Views
Abstract
Mitochondrial dysfunction is a principal feature of acute pancreatitis (AP) although the underlying mechanisms are still unclear. AP precipitants induce Ca2+-dependent formation of the mitochondrial permeability transition pore (MPTP) in pancreatic acinar cells (PACs), leading to ATP depletion and necrosis. Evaluations of mitochondrial bioenergetics have mainly been performed in isolated PACs using confocal microscopy, with assessment of mitochondrial membrane potential, NADH/FAD+ and ATP levels, coupled with patch-clamp electrophysiology. These studies are technically demanding and time-consuming. Application of Seahorse flux analysis now allows detailed investigations of bioenergetics changes to be performed in cell populations using a multi-well plate-reader format; rates of oxygen consumption (OCR) and extracellular acidification (ECAR) provide important information about cellular respiration and glycolysis, respectively. Parameters such as maximal respiration, ATP-linked capacity and proton leak can be derived from application of a respiratory function “stress” test that involves pharmacological manipulation of the electron transport chain. The use of Seahorse Flux analysis therefore provides a quick, and convenient means to measure detailed cellular bioenergetics and allows results to be coupled with other plate-reader based assays, providing a fuller understanding of the pathophysiological consequences of mitochondrial bioenergetics alterations.
Keywords: Mitochondrial dysfunctionBackground
Mitochondrial dysfunction is a core feature of acute pancreatitis (AP), a debilitating and potentially fatal disease for which there is currently no specific therapy (Criddle, 2016; Habtezion et al., 2019). The elucidation of pivotal pathological mechanisms which underlie mitochondrial damage in pancreatic acinar cells (PACs) is paramount for the development of new therapies. Previous evaluations of mitochondrial bioenergetics in isolated PACs have been mostly performed using confocal microscopy, including assessments of mitochondrial membrane potential (tetramethyl rhodamine methyl ester: TMRM), NADH/FAD+ autofluorescence and ATP (Magnesium Green), coupled with patch-clamp electrophysiology (Voronina et al., 2002; Criddle et al., 2004 and 2006). Such studies have pinpointed a reduction of ATP in response to precipitants of AP, via the opening of the mitochondrial permeability transition pore (MPTP), as a critical event that leads to necrotic cell death (Criddle et al., 2006; Mukherjee et al., 2016); supplementation with intracellular ATP ameliorated damage. Furthermore, luciferase measurements in PACs have provided details of changes of both mitochondrial and cytosolic ATP concentrations induced by pathophysiological stimulation (Voronina et al., 2010). Such experimental approaches in single cells, however, are technically difficult and time-consuming. In contrast, population-based assays provide important information about mitochondrial dysfunction and cell death using a convenient plate-reader format (Armstrong et al., 2019). The use of Seahorse Flux analysis allows a detailed evaluation of bioenergetics changes to be performed in PACs, measuring rates of oxygen consumption (OCR) and extracellular acidification (ECAR); these inform about cellular respiration and glycolysis, respectively. A respiratory function “stress” test can further be applied in which pharmacological manipulation of the electron transport chain (ETC) is used to derive parameters such as the Maximal Respiration, Spare Respiratory Capacity, ATP-linked Turnover and Non-mitochondrial Respiration. Such detailed bioenergetics information can be coupled with parallel studies of apoptosis and necrosis to inform the influence of mitochondrial dysfunction on cell death patterns (Armstrong et al., 2018).
Materials and Reagents
- 70 µm filters (Fisherbrand, catalog number: 22363548 )
- Tissue paper (Kimberly-Clark White Roll, Fisher Scientific, catalog number: 12774286 )
- 30 G needle (Microlance 3, 30G x 13 mm) (BD, catalog number: 304000 )
- 1.5 ml microcentrifuge tube (Starlab, catalog number: E1415-1510 )
- Centrifuge tubes
50 ml tube (Starlab, catalog number: E1450-0200 )
15 ml tube (Starlab, catalog number: E1415-0200 ) - Pipette tips
200 µl tip (Starlab, catalog number: S1111-0806 )
1 ml tip (Starlab, catalog number: S1111-6801 ) - Pasteur pipettes 3 ml (Starlab, catalog number: E1414-0311 )
- Male CD1 or C57BL6/J mice, 8-12 weeks old (Charles River)
- Collagenase, Purified (Worthington Biochemical Corporation, catalog number: CLSPA )
- PI-CassetteTM (Chemometec, Nucleocounter, catalog number: 941-0001 )
- Reagent A100; Lysis buffer (SKU: 910-0003, Chemometec, Nucleocounter)
- Reagent B; Stabilizing buffer (SKU: 910-0002, Chemometec, Denmark)
- XF24 Fluxpak, containing cell plates, cartridge consisting of sensor and utility plates and Seahorse XF calibrant (Agilent, Seahorse, catalog number: 100850-100 )
- Matrigel basement membrane matrix (Corning, catalog number: 354234 )
- Dimethyl sulfoxide (DMSO, Sigma, catalog number: D2650 )
- Ethyl alcohol, Pure (Sigma, catalog number: 459836 )
- Oligomycin A (Sigma, Merck, catalog number: 75351 )
- Carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone (FCCP, C2920, Sigma, Merck)
- Antimycin A from Streptomyces sp. (Sigma, Merck, catalog number: A8674 )
- Rotenone (Sigma, Merck, catalog number: R8875 )
- DMEM (Sigma, Merck, catalog number: D5030 )
- L-Glutamine 100x (Gibco, Life Technologies, cataloge number: 25030-081 )
- D-Glucose (Sigma, Merck, catalog number: G7528 )
- Sodium pyruvate (Sigma, Merck, catalog number: P8574 )
- Sodium chloride (Sigma, Merck, catalog number: S9888 )
- Potassium chloride (Sigma, Merck, catalog number: P3911 )
- Magnesium chloride (Sigma, Merck, catalog number: M1028 )
- Calcium chloride (Sigma, Merck, catalog number: 21115 )
- Extracellular solution (see Recipes)
- Seahorse media (see Recipes)
Equipment
- Surgical scissors
- Water bath
- -20 °C freezer
- Multi-channel pipette 20-200 µl
- Seahorse XF24 Extracellular Flux Analyser (Agilent), although other Seahorse Flux Analysers could be used
- Cell Counter NucleoCounter® NC-100TM (900-0004, ChemoMetec, Denmark)
- Inverted light microscope (GX Microscope XDS-3)
Software
- Wave (Seahorse, Agilent. https://www.agilent.com)
- Prism (GraphPad Software Inc., La Jolla, CA. https://www.graphpad.com)
Procedure
Day 1
- Pancreatic acinar cell preparation
- Sacrifice young (8-12 weeks old) adult CD1 or C57BL/6 (wild type) mouse using an approved Schedule 1 procedure.
- Place mouse down on right hand side on fresh tissue paper.
- Using forceps and scissors cut away fur in the side abdominal area. Once removed make a similar removal of lower skin epidermis to expose the abdominal cavity.
- Locate the pancreas below the spleen gently with forceps and cut free with a small pair of surgical scissors.
- Place the freshly excised pancreas in 7 ml ice cold extracellular solution and keep on ice until digestion.
- Pre-warm 1 ml of collagenase solution (200 U/ml) to 37 °C in a water bath.
- Carefully inject the pre-warmed collagenase using a 30 G needle into the pancreas samples placed in a weighing boat until the pancreas inflates. Draw up any remaining collagenase not retained within the pancreas from the weighing boat and re-inject into pancreas.
- Place the inflated pancreas into a 1.5 ml microcentrifuge tube and add any remaining collagenase.
- Place in a 37 °C water bath for 17 min.
Note: This time is dependent on the brand and concentration of collagenase used. You may need to optimize so that you get a optimal amount of cell viability and cell number achieved. - After incubation for 17 min remove from water bath and decant contents of the 1.5 ml microcentrifuge tube into a 15 ml falcon tube labeled ‘Panc’, quickly add 5 ml of extracellular solution.
- Using a 1 ml pipette tip, cut at an angle across the tip, as shown in Figure 1i and the volume set at 700 µl vigorously pipette the whole pancreas tissue up and down, in and out of the pipette tip to allow for tissue dissociation (trituration).
Note: Repeat process until the mixture becomes cloudy with the release of cells.
- Upon cessation of pipetting quickly remove the cellular solution into a fresh 15 ml tube labeled ‘Cells’, using a 3 ml pasteur pipette being careful to leave behind any pieces of pancreas tissue.
- Add 5 ml of extracellular solution to the 15 ml tube ‘Panc’.
- Using a fresh 1 ml pipette tip, cut at a smaller angle than previously used in Step A11 across the pipette tip, as shown in Figure 1ii and the volume set at 700 µl. Vigorously pipette the whole pancreas tissue in and out of the pipette tip to allow further tissue dissociation.
- Upon cessation of pipetting quickly remove the cellular solution into 15 ml tube ‘Cells’, using a 3 ml pasteur pipette and being careful to leave behind any pieces of pancreas tissue.
- Repeat Steps A14-A16 for a third and final time using a fresh 1 ml pipette tip cut at an angle across the tip, as shown in Figure 1iii.
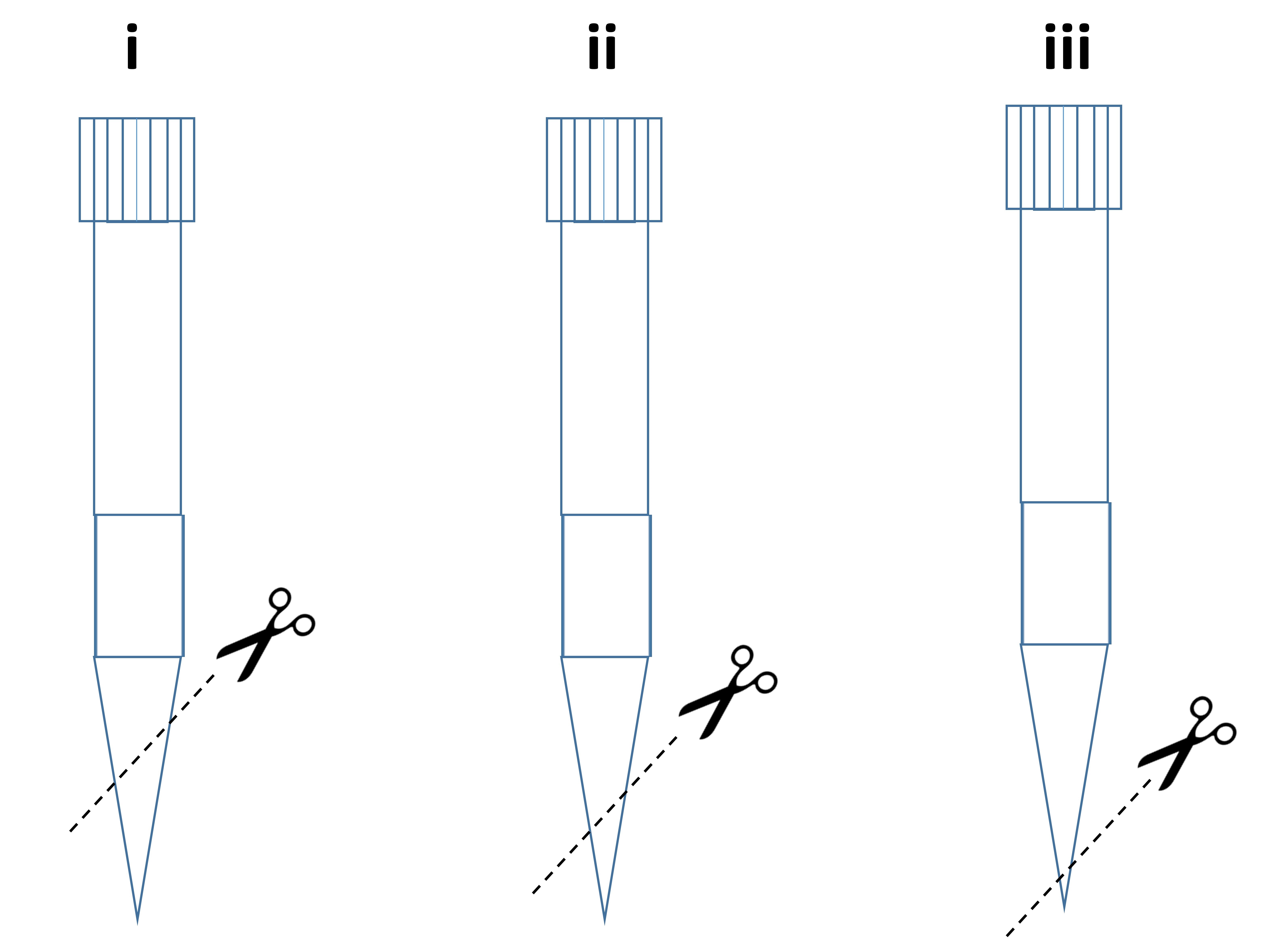
Figure 1. How to prepare 1 ml pipette tips for dissociation - Discard any tissue remaining in a 15 ml tube labeled ‘Panc’.
- Centrifuge 15 ml tube labeled ‘Cells’ for 2 min, 130 x g, room temperature.
- Carefully pour off the supernatant leaving behind the loose cell pellet.
- Resuspend the cell pellet in 1 ml of chilled extracellular solution.
- Add an additional 9 ml of chilled extracellular solution.
- Set up a 70 micron filter in a 50 ml centrifuge tube (labeled and dated) and pre-wet with a small amount of extracellular fluid.
- Filter the 10 ml of cell suspension from the 15 ml tube ‘Cells’ through the filter into the 50 ml tube.
- Rinse the filter and any cells left trapped in the filter through with approximately 30 ml of chilled extracellular fluid until the 50 ml centrifuge tube is filled to the 40 ml mark.
- Centrifuge 50 ml tube labeled ‘Cells’ for 2 min, 130 x g, room temperature.
- Carefully pour off the supernatant leaving behind the loose cell pellet.
- Resuspend the cell pellet in 1 ml of chilled extracellular solution.
- Add an additional 9 ml of chilled extracellular solution.
- Place the 50 ml centrifuge tube with sample overnight at 4 °C.
- Oxygen consumption and lactate production analysis cell plate preparation and cartridge rehydration
- Remove a Seahorse XF 24 well cell plate and remove from packaging.
- Take a pre-aliquotted 250 µl vial of Matrigel from storage in a -80 °C freezer and thaw on ice.
- Adapt Corning’s ‘Thin Coating Method’ for use on a Seahorse XF24 cell plate as follows:
- Using cooled pipettes, mix the BD Matrigel Basement Membrane Matrix to homogeneity.
- Dilute Corning Matrigel Basement Membrane Matrix using serum-free medium Seahorse media.
- Carefully add 36 µl diluted BD Matrigel Basement Membrane Matrix to Seahorse XF 24 well cell plate being sure not to create any bubbles.
- Incubate at room temperature for 30 min.
- Aspirate unbound material, discard and gently rinse by pipetting 100 µl chilled serum-free Seahorse media into each well and then immediately aspirating and discard, using a multichannel pipette.
- Pipette 100 µl chilled serum-free Seahorse media into each well and leave at 4 °C until ready for use.
- Remove a Seahorse XF 24 cartridge from packaging.
- Remove the sensor plate (green upper half of the cartridge) and place lid down on the bench.
- Pipette 1 ml of XF callibrant into each of the 24 wells of the utility plate and return the sensor plate so the probes of each well are submerged in XF callibrant.
- Place in a 37 °C CO2 free incubator overnight until ready for use.
Day 2
- Assess pancreatic cell viability
- The next day the cell suspension is mixed to obtain a homogenous suspension by gently pipetting up and down using a 1 ml pipette.
- Pipette a 1 ml cell sample from the cell suspension into a 1.5 ml microcentrifuge tube.
- Label a PI-CassetteTM ‘NV’ (non-viable).
- Draw the cell suspension into a PI-CassetteTM by inserting the tip of the cassette into the cell suspension in the 1.5 ml microcentrifuge tube and pressing the piston.
- Set aside the ‘NV’ PI-Cassette™ to be analysed.
- Pipette 100 µl of the 1ml cell sample from the 1.5 ml microcentrifuge tube into a second 1.5 ml microcentrifuge tube. Return any cell sample remaining to the original cell suspension 50 ml centrifuge tube ‘Cells’.
- Add 100 µl volume of Reagent A100 to the microcentrifuge tube with the 100 µl cell sample.
- Mix by pipetting.
- Add 100 µl volume of Reagent B to the mixture of cell suspension and Reagent A100.
- Mix by pipetting.
- Label a PI-CassetteTM ‘T’ (total).
- Draw the diluted cell suspension into a PI-CassetteTM by inserting the tip of the cassette into the cell suspension and pressing the piston.
- Immediately place the loaded PI-CassetteTM in the NucleoCounter® NC-100TM sample tray, press RUN.
- After approximately 45 s the total cell concentration (cells/ml) is presented in the bottom right.
- The cell count produced will normally be in the order of x 105. Divide by 10 to achieve x 106 for the following calculations.
- Calculate total cell number and percent viability as follows:
Cell viability = [‘NV’/(‘T’ x 3)] x 100
Total viable cell count = [(‘T’ x 3) – ‘NV’] x volume of cell suspension
Examples of total cell count and percent viability previously achieved when isolating acinar cells from two separate pancreas (taken from two animals) at the same time and pooled for analysis are shown in Table 1.
Table 1. Examples of total cell count and percent viability previously achieved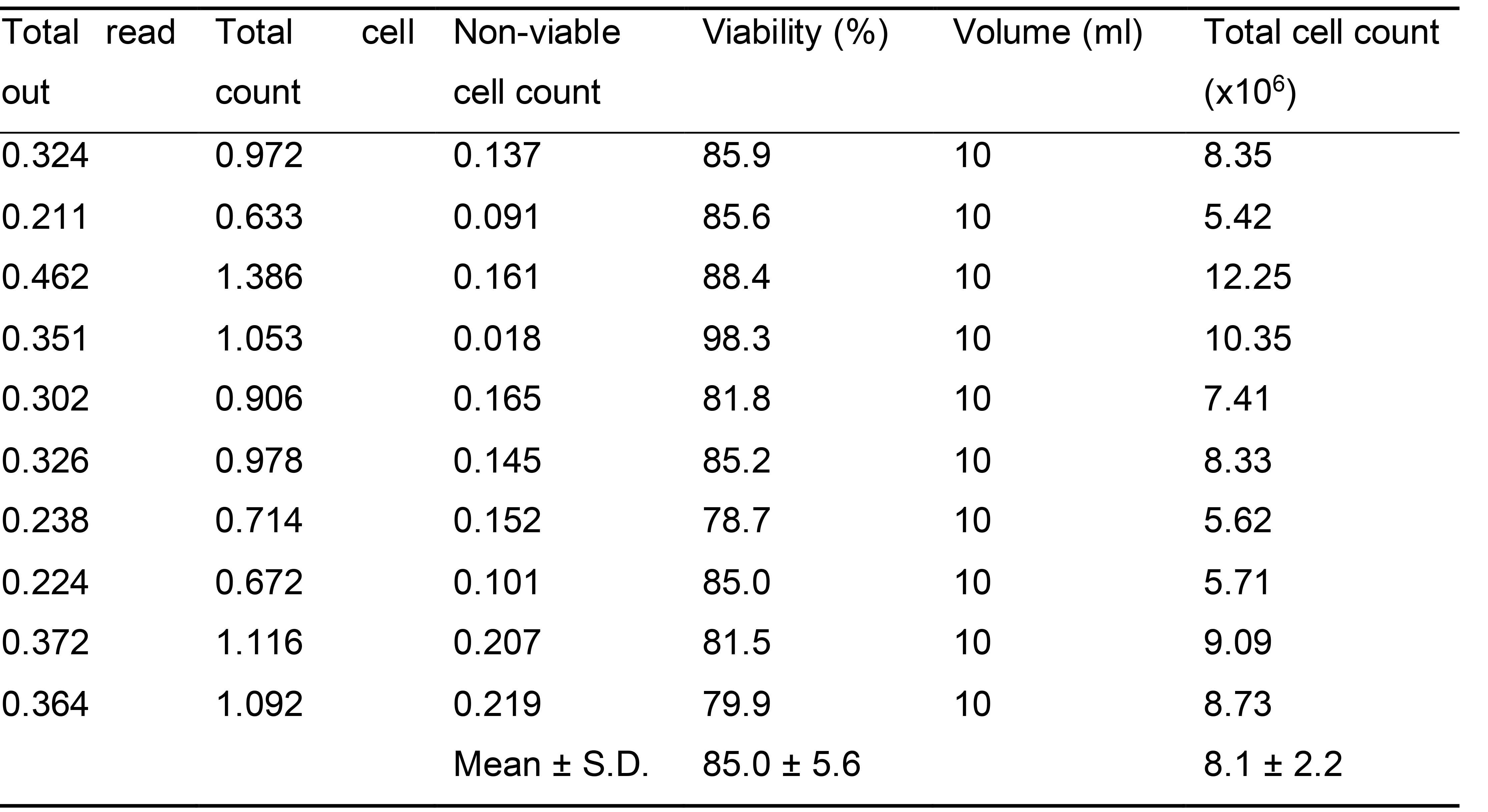
- Centrifuge 50 ml tube labeled ‘Cells’ for 2 min, 130 x g, room temperature.
- Carefully pour off the supernatant leaving behind the loose cell pellet.
- Resuspend the cell pellet with 1 ml of extracellular solution.
- The cell suspension needs to be adjusted to a concentration of 1 x 106/ml. This is achieved by adjusting the volume to the same value as the total number of cells (Total viable cell count = 5 x 106, adjust cell volume to 5 ml). Centrifuge 50 ml tube labeled ‘Cells’ for 2 min, 130 x g, room temperature.
- The cells are now ready to be seeded onto the Matrigel coated Seahorse XF 24 well cell plate (Figure 2).

Figure 2. Well allocation for blank well for XF24 cell plate - Remove the XF 24 well cell plate and remove the 100 µl serum-free Seahorse media from each well.
- Continuously mixing the cell suspension during seeding by pipetting up and down, transfer 75 µl of cell suspension into each well leaving the following cells blank to give a total of 75,000 murine pancreatic cells per well.
- Place the lid back onto the XF24 cell plate and place in a 37 °C CO2 free incubator for 1 h for the cells to adhere.
- Whilst the cells are adhering the XF24 cartridge can be loaded with the compounds that comprise a mitochondrial “stress” test.
- After one hour remove the 75 µl of extracellular solution from the cells. Quickly add 450 µl pre-warmed serum free Seahorse media to every well. Return to a 37 °C CO2 free incubator until ready for use.
- Preparing and loading compounds for the mitochondrial “stress” test
- There are two types of assays that can be performed:
Standard Assay–only involves the injection of modulators included in the kit.
Modified Assay–includes an additional injection of a test compound prior to oligomycin injection, and Port A is used for the testing compound. - Refer to Table 2 for loading volume and port designation for compounds in different types of assays.
Table 2. Loading regime for mitochondrial stress test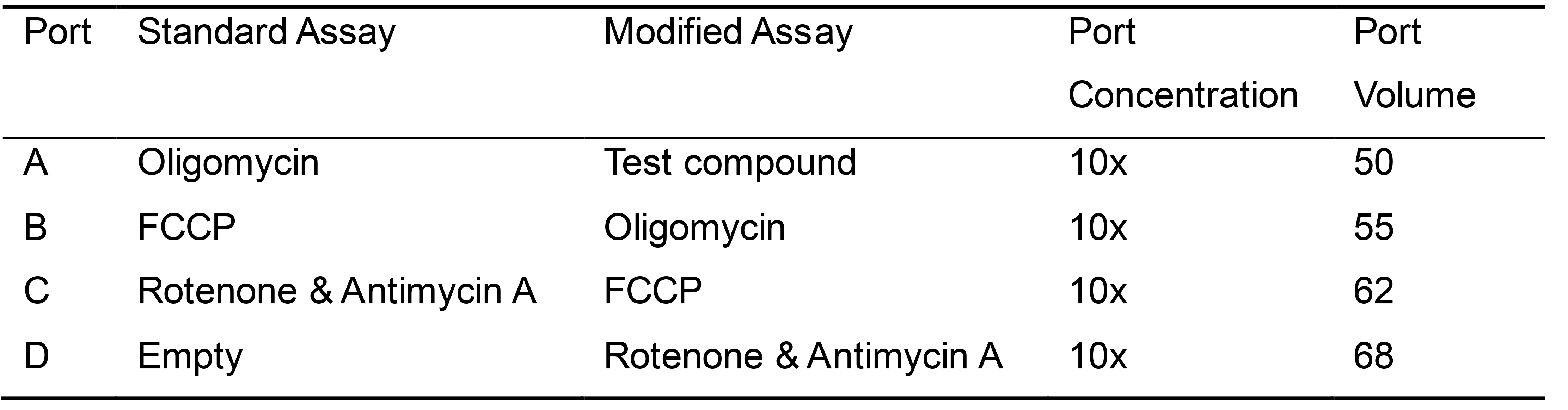
- Make a working concentration of 10x the final required concentration required from stock solutions stored at -20 °C as follows:
- Oligomycin stock solution in DMSO
10 µl in 1 ml of Seahorse media
For a 10x solution of 10 µg/ml for a final concentration of 1 µg/ml. - Carbonyl cyanide-4 (trifluoromethoxy) phenylhydrazone (FCCP) stock solution in DMSO
3 µl in 1 ml of Seahorse media
For a 10x solution of 3 µM for a final concentration of 0.3 µM. - Rotenone stock solution in DMSO and Antimycin A stock solution in ethanol
10 µl of each in 1 ml of Seahorse media
For a 10x solution of 20 µg/ml for a final concentration of 2 µg/ml.
- Oligomycin stock solution in DMSO
- Make a vehicle control solution for any well or port that is not receiving a compound or stress test with appropriate amounts of DMSO/Ethanol or other vehicle utilized for specific compounds.
- Remove both the sensor plate and the corresponding utility plate together from the 37 °C CO2 free incubator and remove the lid.
- Load the corresponding volume of each compound appropriate for the assay type into the appropriate port on the Sensor plate.
- Once all the compounds have been added to the ports, the lid can be replaced on to the Sensor plate and return to a 37 °C CO2 free incubator until ready for use.
- There are two types of assays that can be performed:
- Oxygen consumption and lactate production analysis
- Programme the Seahorse Extracellular Flux instrument as follows:
- Calibration.
- Equilibration.
- [Mix–2 min, Wait–2 min, Measure–2 min] x 5 to ensure a stable baseline.
- Inject Port A.
- [Mix–2 min, Wait–2 min, Measure–2 min] x 3 for standard assay oligomycin injection or x 5 typically for modified assay which gives 30 min for any test compound application.
- Inject Port B.
- [Mix–2 min, Wait–2 min, Measure–2 min] x 3 for standard assay FCCP injection and also x 3 for modified assay, oligomycin injection.
- Inject Port C.
- [Mix–2 min, Wait–2 min, Measure–2 min] x 2 for standard assay Antimycin A/Rotenone injection and x 3 for modified assay, FCCP injection.
- Inject Port D (modified assay only).
- [Mix–2 min, Wait–2 min, Measure–2 min] x 2 for modified assay only, Antimycin A/Rotenone injection.
- After reviewing the group definitions, plate map layout, click Start Run.
- After you enter the save location for your result file (following completion of the assay), the tray door on the XF24 Analyser will open.
- The sensor cartridge fits properly on the Utility plate.
- The lid is removed from the sensor cartridge.
- Proper orientation (direction) of the sensor cartridge on the Utility plate.
- Place the sensor cartridge (hydrated and loaded with compounds) and Utility plate onto the tray when prompted.
- Press “Start” to initiate sensor cartridge calibration.
- Time to complete calibration is approximately 10-20 min (for assays at 37 °C). For XF assays performed at temperatures other 37 °C an additional 30 min of pre-calibration time will be added to ensure accurate data acquisition.
- Once sensor cartridge calibration is complete, the instrument controller will display the Load Cell Plate dialogue.
- Click Open Tray to eject the Utility plate and load the Cell Plate on the tray. The sensor cartridge remains inside the XF Analyser for this step.
Important! Before loading the Cell plate, ensure:- The lid is removed the Cell Plate.
- Proper orientation (direction) of the Cell Plate on the tray.
- After placing the Cell Plate on the tray, click Load Cell Plate to initiate equilibration.
- After completing equilibration, the assay will automatically begin acquiring baseline measurements (as outlined in your instrument protocol).
- Once the final measurement command in the instrument protocol is completed, Wave Controller software will display the Unload Sensor Cartridge dialog.
- Click Eject when ready to eject the sensor cartridge and cell plate. Set aside for later analysis if necessary (example - cell count normalization).
- After removing the sensor cartridge and cell plate, the Assay Complete dialogue will appear.
- Click View Results to immediately open your assay result file. Download both files for the experiment.
- Excel file.
- Xfd. File.
- Programme the Seahorse Extracellular Flux instrument as follows:
Data analysis
- Open the Xfd file in Wave software.
- You need to change the file format into a Prism file by selecting ‘save as’ and then choosing the .pzfx format.
- The data can now be opened directly in prism.
- Each experimental condition needs at least 3 wells per plate and for statistics needs at least n = 6 plates using fresh pancreas isolated from a separate mouse each time.
- Exclusion criteria:
- Unstable baseline, usually when the baseline shows a steady rate of decline excessively low or high baseline < 200 or > 800 pMol/min OCR.
- Lack of response by stress test control wells to stress test compounds.
- Outlier data points–entire series for a well and/or individual data points.
Notes
Important! Before you start your XF Assay
- Visually inspect the injection ports for even loading. The liquid should be in the port, make sure there are no residual drops on the top of the sensor cartridge.
- View cells under a microscope to:
- Confirm cell health, morphology, seeding uniformity and purity (no contamination).
- For adherent cells, ensure cells are adhered with a consistent monolayer and were not washed away during washing step.
Recipes
- Extracellular solution [(mM) to make a 500 ml solution, Table 3]
Table 3. Recipe for Extracellular Solution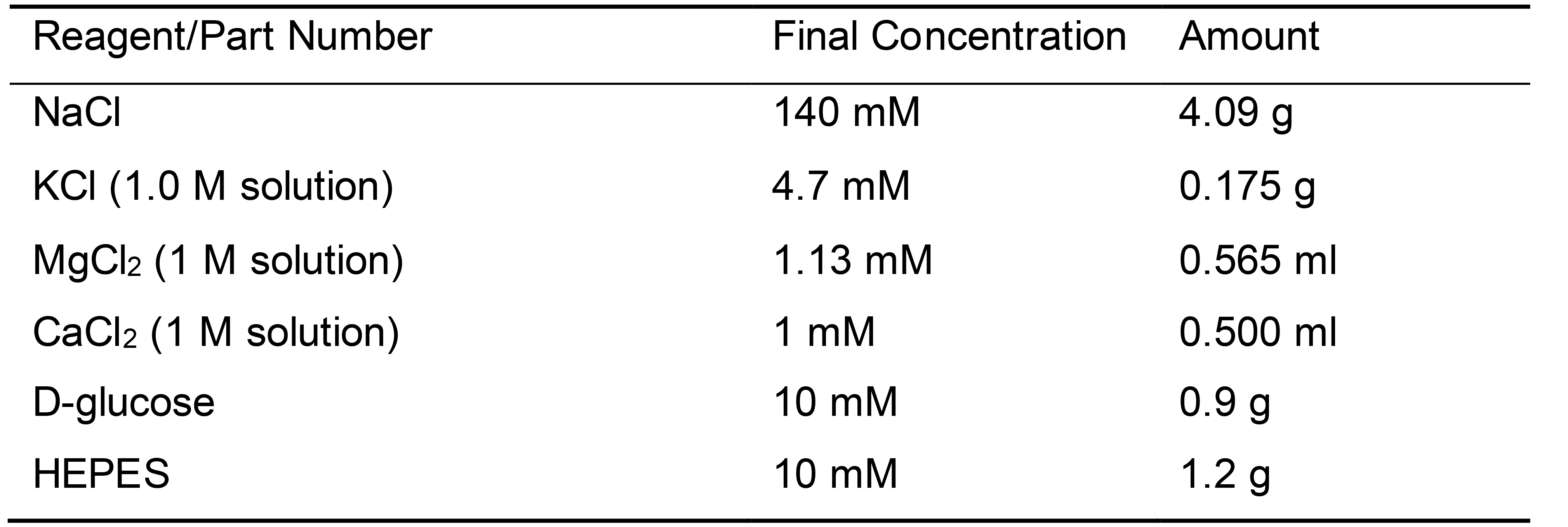
(adjust to pH 7.25 using NaOH) - Seahorse media
- Prepare DMEM by adding 800 ml dH2O to powdered DMEM
- Prepare assay medium by supplementing DMEM medium with 1 mM pyruvate, 2 mM glutamine, and 10 mM glucose as in the table below (Table 4):
Table 4. Recipe for serum free non-buffered Seahorse media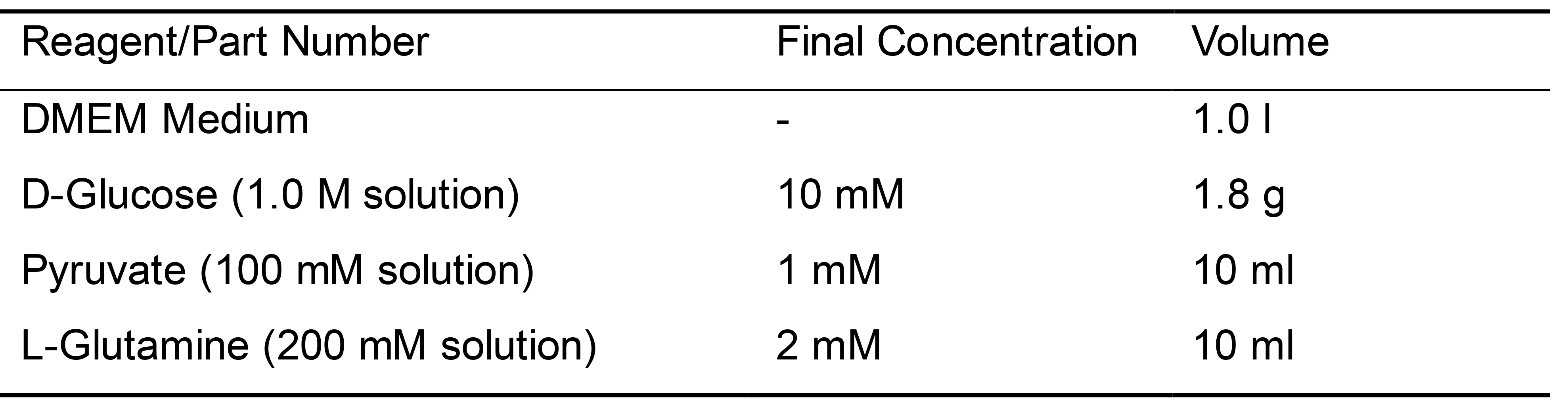
- Bring XF medium with supplements to pH 7.4 and adjust the final volume to 1 L, transfer into a Class 2 microbiological safety cabinet
- Filter with a filter pipette into sterile 50 ml aliquot centrifuge tubes. Store in a fridge for up to 1 month
Acknowledgments
This work was supported by the Wellcome Trust (102381/Z/13/Z), the Medical Research Council (UK) and by the National Institute for Health Research (UK) grant to the NIHR Liverpool Pancreas Biomedical Research Unit. This protocol was first published in the original article by Armstrong et al. (2018).
Competing interests
The authors declare that they have no conflicts of interest with the contents of this article.
Ethics
The animals were humanely sacrificed by increasing CO2 (schedule 1 procedure) in accordance with the Animals (Scientific Procedures) Act (1986) under Establishment License 40/2408 with approval by the University of Liverpool Animal Welfare Committee and Ethical Review Body (X70548BEB and PPL 70/8109).
References
- Armstrong, J. A., Cash, N. J., Morton, J. C., Tepikin, A. V., Sutton, R. and Criddle, D. N. (2019). Mitochondrial targeting of antioxidants alters pancreatic acinar cell bioenergetics and determines cell fate. Int J Mol Sci 20(7): 1700.
- Armstrong, J. A., Cash, N. J., Ouyang, Y., Morton, J. C., Chvanov, M., Latawiec, D., Awais, M., Tepikin, A. V., Sutton, R. and Criddle, D. N. (2018). Oxidative stress alters mitochondrial bioenergetics and modifies pancreatic cell death independently of cyclophilin D, resulting in an apoptosis-to-necrosis shift. J Biol Chem 293(21): 8032-8047.
- Criddle, D. N. (2016). Reactive oxygen species, Ca2+ stores and acute pancreatitis; a step closer to therapy? Cell Calcium 60(3): 180-189.
- Criddle, D. N., Murphy, J., Fistetto, G., Barrow, S., Tepikin, A. V., Neoptolemos, J. P., Sutton, R. and Petersen, O. H. (2006). Fatty acid ethyl esters cause pancreatic calcium toxicity via inositol trisphosphate receptors and loss of ATP synthesis. Gastroenterology 130(3): 781-793.
- Criddle, D. N., Raraty, M. G., Neoptolemos, J. P., Tepikin, A. V., Petersen, O. H. and Sutton, R. (2004). Ethanol toxicity in pancreatic acinar cells: mediation by nonoxidative fatty acid metabolites. Proc Natl Acad Sci U S A 101(29): 10738-10743.
- Habtezion, A., Gukovskaya, A. S. and Pandol, S. J. (2019). Acute pancreatitis: a multifaceted set of organelle and cellularinteractions. Gastroenterology 156(7): 1941-1950.
- Mukherjee, R., Mareninova, O. A., Odinokova, I. V., Huang, W., Murphy, J., Chvanov, M., Javed, M. A., Wen, L., Booth, D. M., Cane, M. C., Awais, M., Gavillet, B., Pruss, R. M., Schaller, S., Molkentin, J. D., Tepikin, A. V., Petersen, O. H., Pandol, S. J., Gukovsky, I., Criddle, D. N., Gukovskaya, A. S., Sutton, R. and Unit, N. P. B. R. (2016). Mechanism of mitochondrial permeability transition pore induction and damage in the pancreas: inhibition prevents acute pancreatitis by protecting production of ATP. Gut 65(8): 1333-1346.
- Voronina, S., Sukhomlin, T., Johnson, P. R., Erdemli, G., Petersen, O. H. and Tepikin, A. (2002). Correlation of NADH and Ca2+ signals in mouse pancreatic acinar cells. J Physiol 539(Pt 1): 41-52.
- Voronina, S. G., Barrow, S. L., Simpson, A. W., Gerasimenko, O. V., da Silva Xavier, G., Rutter, G. A., Petersen, O. H. and Tepikin, A. V. (2010). Dynamic changes in cytosolic and mitochondrial ATP levels in pancreatic acinar cells. Gastroenterology 138(5): 1976-1987.
Article Information
Copyright
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Armstrong, J. A., Sutton, R. and Criddle, D. N. (2020). Pancreatic Acinar Cell Preparation for Oxygen Consumption and Lactate Production Analysis. Bio-protocol 10(10): e3627. DOI: 10.21769/BioProtoc.3627.
- Armstrong, J. A., Cash, N. J., Ouyang, Y., Morton, J. C., Chvanov, M., Latawiec, D., Awais, M., Tepikin, A. V., Sutton, R. and Criddle, D. N. (2018). Oxidative stress alters mitochondrial bioenergetics and modifies pancreatic cell death independently of cyclophilin D, resulting in an apoptosis-to-necrosis shift. J Biol Chem 293(21): 8032-8047.
Category
Cell Biology > Cell metabolism > Respirometry
Biochemistry > Other compound > Acid
Biochemistry > Other compound > Oxygen
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









