- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Generation, Analyzing and in-vivo Drug Treatment of Drosophila Models with IBMPFD
Published: Vol 10, Iss 10, May 20, 2020 DOI: 10.21769/BioProtoc.3621 Views: 5203
Reviewed by: Nicoletta CordaniSalma MerchantAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Bimolecular Fluorescence Complementation (BiFC) for Studying Sarcomeric Protein Interactions in Drosophila
Océane Marescal [...] Nicanor González-Morales
Apr 5, 2020 6697 Views

Identification and Monitoring of Nucleotide Repeat Expansions Using Southern Blotting in Drosophila Models of C9orf72 Motor Neuron Disease and Frontotemporal Dementia
Joanne L. Sharpe [...] Ryan J. H. West
May 20, 2022 2364 Views

Detecting Native Protein–Protein Interactions by APEX2 Proximity Labeling in Drosophila Tissues
Jhen-Wei Wu [...] Yu-Chiuan Chang
Oct 20, 2024 2147 Views
Abstract
Missense mutations of p97/cdc48/Valosin-containing protein (VCP) cause inclusion body myopathy, Paget disease with frontotemporal dementia (IBMPFD) and other neurodegenerative diseases. The pathological mechanism of IBMPFD is not clear and there is no treatment. We generated Drosophila models of IBMPFD in adult flight muscle in vivo. Here we describe a variety of assays to characterize disease pathology and dissect disease mechanism, and the consequences of in vivo feeding of VCP inhibitors.
Keywords: DrosophilaBackground
Mutations of VCP/p97 cause inclusion body myopathy, Paget disease of the bone and frontotemporal dementia (IBMPFD), a degenerative disease in multiple systems including the brain, muscles and bones in an autosomal dominant fashion (Watts et al., 2004). Mutations in VCP are also associated with 1-2% of cases of sporadic amyotrophic lateral sclerosis (ALS), as well as hereditary spastic paraplegia and Charcot-Marie-Tooth 2 neuropathy (Abramzon et al., 2012; de Bot et al., 2012; Gonzalez et al., 2014). The R155H mutation is the most frequently identified in patients, while individuals with the A232E mutation have the most severe clinical manifestation (Kimonis et al., 2008a; Ritson et al., 2010). 90% of IBMPFD patients display myopathy, frequently the earliest symptom (Weihl et al., 2009). 50% of patients will develop Paget’s disease of bone, affecting skull, spine, hips and long bones. One-third of the patients develop frontotemporal dementia (Kimonis et al., 2008b; Weihl et al., 2009). Patients ultimately develop cardiopulmonary failure (Kimonis et al., 2008b; Weihl et al., 2009). VCP encodes a highly conserved and abundant AAA+ ATPase which participates in multiple cellular processes (Meyer et al., 2012). Since VCP assembles as a hexamer, it has been controversial whether disease mutants with increased ATPase activity cause disease through a dominant-active (Chang et al., 2011) or dominant-negative mechanism (Ju et al., 2009; Ritz et al., 2011; Bartolome et al., 2013; Kim et al., 2013; Kimura et al., 2013). We built in vivo IBMPFD models to understand the pathogenesis of the disease and find potential treatments.
The specific assays for IBMPFD disease in flies include in situ cell death detection, muscle protein Western blot assays, immunofluorescence staining of disease markers, muscle integrity assay/Toluidine Blue staining, and muscle mitochondrial ultrastructural studies/Electron Microscopy (EM) imaging, and in vivo VCP inhibitors treatment (Yun et al., 2008; Zhang et al., 2017; Ma et al., 2018).
Materials and Reagents
- Razor blade (VWR North American, catalog number: 55411-050 )
- Microcentrifudge tube (Denville Scientific Inc., catalog number: C2171 )
- PCR tube (USA Scientific)
- FORMVAR FILM on 100 Square Mesh Copper Grid (Electronic Microscopy Sciences, catalog number: FF100-Cu )
- FORMVAR CARBON FILM on 2 x 1 mm oval slot Copper Grid (Electronic Microscopy Sciences, catalog number: FCF2010-Cu )
- Disposable pellet pestle (Kimble, catalog number: 749521-0500 )
- PCR tube
- Dissection dish
- Needles
- Pipette tips
- Toothpick (Fisher Scientific Education, S04180 )
- 0.22 µm filter (Millipore, MillexGP Filter Unit)
- Aluminum foil (Fisher Scientific, catalog number: 01-213-101 )
- Immobilon-P PVDF transfer membrane (Millipore, catalog number: IPVH00010 )
- Drosophila strains: UASt-VCP WT, VCP RH and AE lines were gifts from Dr. Tzu Kang Sang (Chang et al., 2011)
- Drosophila Strain
IFM-Gal4, with Gal4 under the control of the indirect flight muscle promoter, derived from the flightin gene (Yun et al., 2014). The IFM promoter provides a strong pulse of expression in late pupal stages and the first few days of adulthood, and is thereafter silent (Kandul et al., 2016). Drosophila strains were maintained in a 25 °C humidified incubator. - 20% paraformaldehyde, EM Grade (Electronic Microscopy Sciences, catalog number: 15713S )
- 8% glutaraldehyde solution, EM Grade (Electronic Microscopy Sciences, catalog number: 16020 )
- 16% paraformaldehyde aqueous solution, EM Grade (Electronic Microscopy Sciences, catalog number: 15710 )
- Schneider’s Buffer (GIBCO, catalog number: 21720-024 )
- In Situ Cell Death Detection Kit (Roche, catalog number: 11684795910 )
- Rhodamine Phalloidin (Life Technology, catalog number: R415 )
- Bovine Serum Albumin (Fisher Bio Reagent, catalog number: BP1600-100 )
- Fluoromount-G (Southern Biotech, catalog number: 0100-01 )
- Pierce RIPA lysate Buffer (Thermo Scientific, catalog number: 89900 )
- Pierce Protease Inhibitor Tablets EDTA free (Thermo Scientific, catalog number: 88266 )
- Laemmli SDS Sample Buffer, 6x (Bioland Scientific LLC, catalog number: SAB02-02 )
- Anti-mouse IgG horseradish peroxidase linked whole antibody (from sheep) (GE Healthcare, catalog number: NXA931 )
- Anti-rabbit IgG horseradish peroxidase linked F(ab’)2 fragments (from donkey) (GE Healthcare catalog number: NA9340V )
- Goat anti-rabbit/mouse Alexa Fluor 546/488 secondary antibody (Life Technologies, catalog numbers: A11034 / A11029 / A11035 / A10036 )
- Tween-20 500 ml (Hoefer, catalog number: 56-40-6 )
- Immobilon Western Chemiluminescent HRP Substrate (Millipore, catalog number: WBKLS0500 ).
- Embed812 (Electronic Microscopy Sciences, catalog number: 14900 )
- Osmium Teroxide (OsO4) 4% aqueous solution (Electronic Microscopy Sciences, catalog number: 19150 )
- DDSA (dodecenyl succinic anhydride) (Electronic Microscopy Sciences, catalog number: 13710 )
- NMA (nadic methyl anhydrate) (Electronic Microscopy Sciences, catalog number: 19000 )
- BDMA (benzyldimethylamine) (Electronic Microscopy Sciences, catalog number: 11400 )
- Propylene Oxide (Electronic Microscopy Sciences, catalog number: 20401 )
- Toluidine Blue O Powder (Electronic Microscopy Sciences, catalog number: 22050 )
- Sodium Borate (Fisher Scientific, catalog number: S249-500 )
- Uranyl Acetate Dihydrate (Ted Pella, catalog number: 19481 )
- Lead Nitrate (Sigma-Aldrich, catalog number: 22862-100G )
- Sodium Citrate, dihydrate (EMD, catalog number: SX0442-1 )
- Sodium Hydroxide Certified ACS Pellets NaOH (Fisher Scientific, catalog number: S318-500 )
- NMS-873 (3-[3-(cyclopentylthio)-5-[[[2-methyl-4'-(methylsulfonyl) [1,1'-biphenyl]-4-yl] oxy] methyl]-4H-1,2,4-triazol-4-yl]-pyridine (Selleckchem, catalog number: S7285 )
- ML240,2-(2-Amino-1H-benzimidazol-1-yl)-8-methoxy-N-(phenylmethyl)-4-quinazolinamine (Sigma-Aldrich, catalog number: 1346527-98-7 )
- Permount mounting medium (Fisher Scientific, catalog number: SP15-100 )
- 0.1% Triton X-100
- PBS
- TUNEL Enzyme
- β-mercaptoethanol
- Ethanol
- Muscle Dissection Fixative Buffer (see Recipes)
- Dissection Buffer (see Recipes)
- TUNEL Blocking Buffer (see Recipes)
- 0.2 M Phosphate Buffer (pH = 7.4) (see Recipes)
- 10 ml EM Fixative Buffer (see Recipes)
- Epon Mix (Medium) (see Recipes)
- 1% Toluidine Blue Staining Solution (see Recipes)
- Uranium Acetate Staining Solution (see Recipes)
- Lead citrate Staining Solution (see Recipes)
Equipment
- Pipette (Denville Scientific Inc., models: P10, P20, P200, P1000)
- Diamond knife Ultra 45° (Diatome, catalog number: MX5341 )
- PELCO Reverse self-closing Tweezers (Ted Pella, catalog numbers: 5373/ 5375-NM )
- DUMONT Biology Grade Tweezers (Ted Pella, catalog numbers: 505/505-U )
- Pellet pestle motor (Kimble/Kontes, catalog number: 749540-0000 )
- JEOL 100CX transmission electron microscope (UCLA Brain Research Institute, Electronic Microscopic Core Facility)
- Ultracut ultramicrotome (Leica EM UC6, Dr. Frank Laski, UCLA Molecular Cellular and Development Biology)
- PELCO Pro Reverse (self-closing) tweezers (Ted Pella, catalog number: 5375-NM )
- Dissection tweezers (Dumont Biology Switzerland, Electronic Microscopic Science)
- Diamond knife (DiATOME, Ultra 45° MX5341 )
- Formvar mesh/slot grid (Electronic Microscopic Science, catalog numbers: FCF2010-Cu , FF100-Cu -50)
- Water bath (Fisher Scientific, model: Isotemp )
- The Mini-PROTEAN® tetra handcast systems and tetra blotting bodule (Bio-Rad)
- Safety fume hood (UCLA facility 4315-3309-2)
- Heat block (Fisher Scientific, model: Isotemp )
Procedure
- Indirect flight muscle dissection
- Anesthetize flies on a CO2 plate. Cut the head and abdomen off using a razor blade.
- Fix the thoraces in 4% paraformaldehyde/Schneider’s Buffer for 45 min at room temperature in either a microcentrifuge tube or PCR tube.
- Transfer the post-fix thoraces into a dissection dish containing 0.1% Triton X-100/PBS. Triton X-100 helps to break the surface tension over the cuticles and facilitates muscle piece dissection in the following steps.
- Indirect flight muscles (IFM, indicated by red arrows in Figures 1A and 1B) are located in the middle of the thoraces. IFMs can be dissected out using either 30-gauge needles or tweezers. Use needles or tweezer to gently open up the thorax from either the dorsal or ventral midline and expose the IFM. The IFMs are two large groups of muscles that run anterior-posterior in the middle of the thorax (Figure 1C). Each group of IFMs contains 5 large muscles that lie parallel to each other and are located at the dorsal surface of the half-dissected thorax. Carefully isolate each muscle by cutting at the ends that attach the muscle to the cuticle. One well-fixed wildtype thorax can generate 5-10 fragments of intact muscle (Figure 1C). Flies that carry an IFM targeted GFP marker can be used to further confirm that the correct muscle has been dissected (Figure 1C).
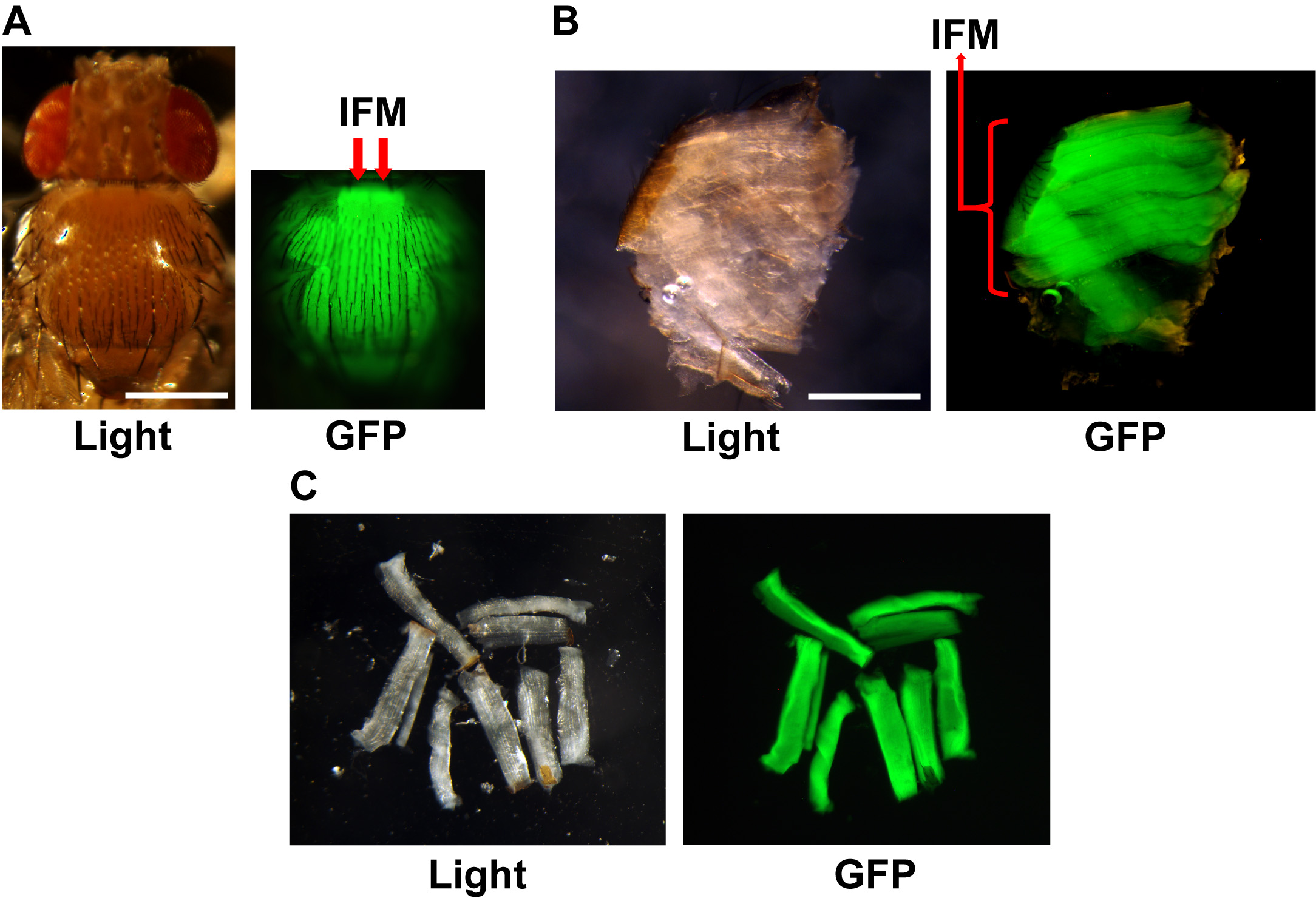
Figure 1. Indirect Flight Muscle (IFM) Dissection. A. The location of IFM in intact thorax (red arrows). B. The sagittal plane of a fixed thorax, red arrow indicating where the IFM are located. C. Dissected muscle pieces in the dissection dish. The genotype of the flies used is IFM-Gal4>UASt-mitoGFP. Scale bar: 0.5 mm.
- In situ cell death detection/TUNEL Assay for IBMPFD flies
- Fix the thoraces and dissect the IFMs from WT, VCP WT, RH and AE flies as described above and collect into PCR tubes.
- Introduce TUNEL Blocking buffer into the tubes containing muscle for 30 min. The TUNEL Blocking buffer contains 0.2% Triton X-100 that permeabilizes and blocks the muscle sample at the same time.
- Add TUNEL Enzyme (4 µl) and 10x Reaction Buffer (36 µl) from the in-situ Cell Death Detection Kit and incubate for 2-3 h at 37 °C in a water bath. Use a pipette to gently mix the sample every 30 min.
- Wash the samples with PBS twice.
- Replace PBS with Fluoromount-G mounting solution. Mount IFMs on a microscopic slide using either a glass pipette or 200 µl pipette tip and visualize the muscle with a confocal microscope using an excitation wavelength of 546 nm. In 6 days old flies, VCP RH and AE expression leads to significant cell death (Zhang et al., 2017). This is observed as extensive red nuclear staining.
- Immunofluorescence staining and confocal microscopy imaging
- Dissect the fixed muscle as described above. Then wash with PBS and permeabilize muscles with 0.2% Triton X-100/PBS for 3-4 h on the rocker at room temperature followed by 5 min wash with 1x PBS.
- For myofibril staining, add Rhodamine Phalloidin (1:500 dilution) and stain for 2 h at room temperature or 4 °C overnight.
- For immunofluorescence staining, fixed muscle fragments are incubated with primary antibody diluted to the desired concentration (1:100 or 1:200) in 0.2% Triton X-100/PBS at 4 °C overnight. We used anti-TARTAR Binding Protein or anti-TDP43 at a concentration of 1:100.
- Wash 3x each for 10 min with 0.2% Triton X-100/PBS.
- Incubate muscle fragments with a goat anti-rabbit/mouse Alexa Fluor 488 or 546 secondary antibody (1:200 in 0.2% Triton X-100/PBS) for 2 h at room temperature or 4 °C overnight.
- Wash three times (10 min each time) with 0.2% Triton X-100/PBS and mount samples on a microscope slide with Fluoromount-G. Samples are visualized under a confocal microscope with an excitation wavelength at 488 nm or 546 nm corresponding to the secondary antibodies. In 6-day old flies, TDP43 localizes both in the nucleus and sarcoplasmic/cytosolic area of the IFM in wildtype flies; the nucleus localization pattern is lost and more aggregates are observed in the sarcoplasmic/cytosolic region in VCP RH and AE mutants (Zhang et al., 2017).
- Protein lysates and Western blot
- Anesthetize flies with CO2, isolate thoraces as above, and put them in a microcentrifuge tube on ice.
Note: The thorax is not fixed. - Add RIPA lysis buffer with protease inhibitors, 10-15 µl for each thorax, 5-10 thoraces for each genotype.
- Homogenize the thoraces in the lysis buffer with prechilled the polypropylene pellet pestles 5-6 times, followed by spinning the pestle with the mortar at the highest speed for 10-15 s. Incubate lysate on ice for 30 min to fully lyse the tissue.
- Centrifuge protein lysates at 10,000 x g for 15 min and transfer the supernatant to a clean microcentrifuge tube.
- Boil the supernatant with 6x SDS sample buffer with β-mercaptoethanol at 95 °C for 5 min.
- Load proteins and let them separate in 8% SDS-PAGE Gels in Tris/SDS running buffer and run it at 80 volts.
- Transfer proteins to PVDF membrane for 2 h in Tris/Glycine Buffer with 15% methanol.
- Incubate transfer membrane with 3% BSA/PBS for 1 h.
- Incubate the membrane with primary antibody diluted in 1% BSA/0.01% Tween-PBS at 4 °C overnight.
- Wash the membrane in 0.01% Tween-PBS for three times (10 min each) and incubate with anti-mouse/rabbit IgG HRP linked secondary antibodies diluted in 5% non-fat milk/0.01% Tween-PBS for 2 h at room temperature.
- Develop the membrane with Immobilon Western Chemiluminescent HRP Substrate Kit. We expressed VCP WT, VCP RH and AE at comparable levels in the thoraces (Zhang et al., 2017).
- Anesthetize flies with CO2, isolate thoraces as above, and put them in a microcentrifuge tube on ice.
- Toluidine blue staining and electronic microscopy for IBMPFD flies
Fixation and Embedding
Note: Perform Step E4 and subsequent steps in a safety fume hood.- Anesthetize flies and cut the thoraces, quickly dip in 95% ethanol (this helps to break the surface tension so as to facilitate subsequent immersion in the EM fixative buffer). Then transfer thoraces to 0.5 ml ice-old EM Fixative Buffer (below) in a microcentrifuge tube.
- Fix for a minimum of 2 h on ice with rocking. If fixation is carried out for longer or the protocol needs to be paused, samples may be left in fixative after the 2 h time point and placed at 4 °C overnight or until ready to proceed. Thoraces will sink to the bottom of microcentrifuge tube.
- Rinse each sample with 0.1 M phosphate buffer for three times (10 min each time) at room temperature.
Note: All subsequent steps should be carried out in a Bio-safety hood. - Post-Fix samples in 1% OsO4 in ddH2O (freshly prepared) for 2 h at room temperature. Samples should not be fixed with OsO4 for more than 2 h or the samples will become brittle.
- Rinse sample with ddH2O for three times (10 min each) at room temperature.
- Dehydrate samples in 70% and 95% ethanol for 5 min each at room temperature.
- Dehydrate samples in 100% ethanol twice (10 min each) at room temperature.
- Dehydrate samples in 100% propylene oxide twice (7 min each) at room temperature.
- Infiltrate samples in 1:1 mixture of 100% propylene oxide and Epon mix for 30 min at room temperature.
- Incubate samples in 100% Epon Mix (1.5% BDMA added) overnight at room temperature.
- Introduce samples in 100% Epon Mix (1.5% BDMA added) into an embedding mold. In the bottom right of each well, put a piece of paper with sample name/number (Figure 2A).
- Polymerize Epon at 60-70 °C overnight.
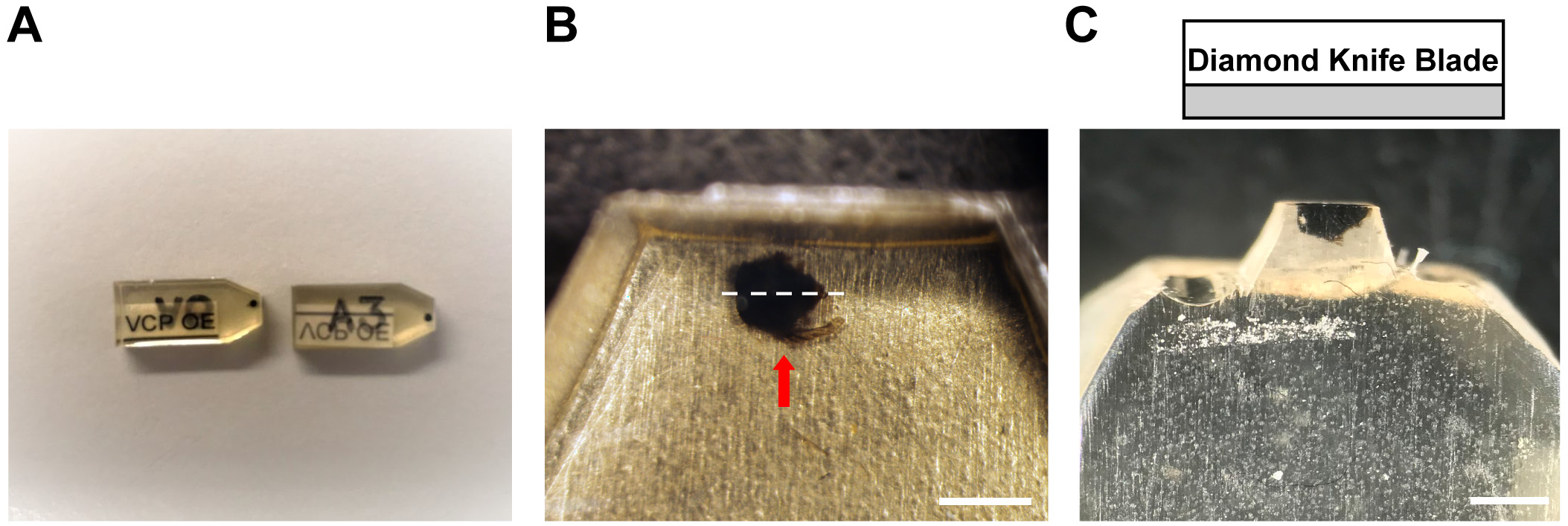
Figure 2. Drosophila thorax embedding and section position. A. Two individual samples embedded and blocked with labels. B. Properly embedded fly thorax (red arrow) in the Epon Resin. The sagittal plane of the thorax (white dotted lines) should be parallel to the edge of the resin block so as to facilitate the section. C. A well-trimmed resin block with a thorax sample after thick section. Glass knife or Diamond knife blades should run parallel to the sagittal plane of the thorax. Scale bar: 1 mm.
- Toluidine Blue Staining
- Well-embedded thorax samples are positioned in the resin block as shown (Figure 2B). The sagittal midline of the thorax is parallel to the edge of the resin block.
- Use a razor blade to cut away the Epon around the sample as much as possible (Figure 2C). Decreasing the section surface will increase the section quality.
- Cut thick sections (1.5-2 µm) for Toluidine Blue staining and to determine the area within which thin sections will be taken for electron microscopy imaging.
- Use a microtome with glass knife to cut thick sections. Transfer sections to water drops on the glass slide using a toothpick.
- Evaporate the water by putting the slide on a 70 °C heat block. This will flatten the section samples.
- Apply 1% Toluidine Blue to fully cover the section sample area and put the slide on the 70 °C heat block until the edge of the Toluidine Blue liquid starts to dry (2-5 min). Do not over dry the Toluidine Blue solution or the section will become contaminated by the dry solute in the staining buffer, which results in degradation of image quality.
- Rinse the slide in ddH2O until non-bound stain is removed.
- Let the section dry completely before applying the Permount mounting solution.
- A well-aligned thick section sample of a wildtype thorax is shown in Figure 3A (Indirect flight muscle pieces can be easily observed). Higher magnification images of the tissue samples can be utilized (Figure 3B) to assay muscle tissue integrity. In 6-day old flies, VCP RH and AE expressing flies have disrupted muscle tissue integrity as compared to WT and VCP WT controls (Zhang et al., 2017).
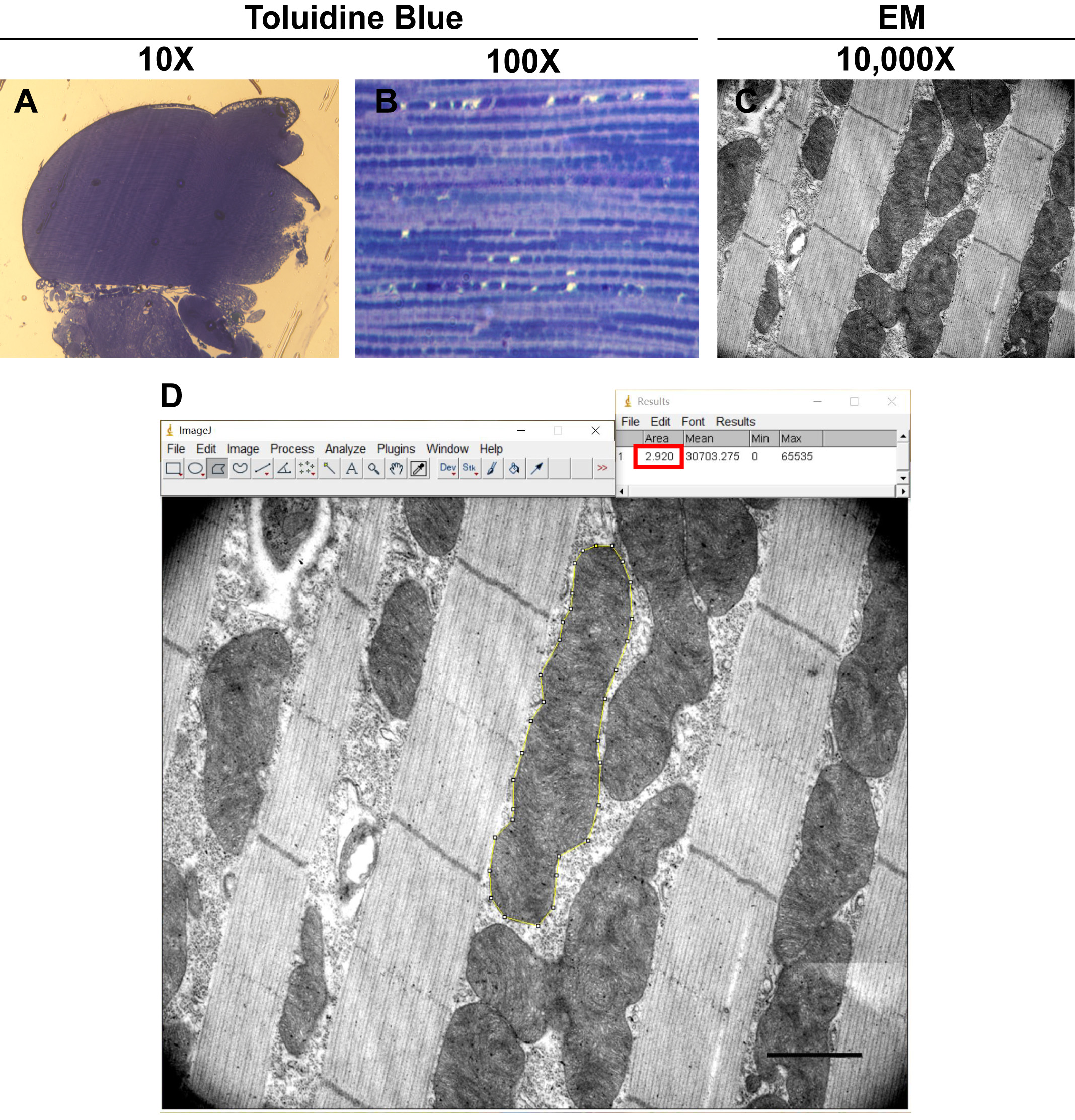
Figure 3. Toluidine Blue and EM images of IFM. A-B. Toluidine Blue staining of thick sections of thorax at different magnifications. C. EM image of a thin section of wildtype thorax. D. Mitochondrial size quantification using ImageJ. Scale bar: 1 µm (119 pixels). Cross-section area of selected mitochondrion is 2.92 µm2 (Red square).
- Electronic microscopy sample preparation
- Select well-aligned samples from the Toluidine Blue staining to be used in thin sectioning (80-90 nm).
- Use a microtome with a Diamond knife (with sterilized ddH2O) to cut thin sections. The sections should appear silver to gold color.
- Use a fine needle to position 1-4 sections in a line on the water surface for grid mounting.
- Pick up an empty slot grid that is not coated with formvar with tweezers. Gently touch the grid to sections floating on the surface. The sections will attach to the middle of the slot within the water drop.
- Use a pair of self-closing tweezers to hold a new formvar-sealed slot grid or mesh grid with the shiny-side up. Place the section-attached slot grid onto the shiny side of the new formvar grid.
- Remove the water drop between two grids by placing the sharp corner of a triangle-shaped filter paper at the edge between the grids. Sections will attach to the formvar-sealed new grids and flatten.
- Remove the empty grid and reuse it to pick up additional sections. The section-attached grid can be stored in the grid box for further staining as described below.
- Prepare the uranium acetate (UA) solution and filter through a 0.22 µm filter system. Float the section side of the grid on the UA solution for 15 min at room temperature. Wash the grid five times (1 min each) in a ddH2O water drop or three times (1 min each) in 3 separate beakers containing ddH2O.
- Put the grid section with shiny side down on a drop of the lead citrate solution and stain for 5-10 min at room temperature. Wear a mask to prevent breathing on to the sections as excess CO2 will affect staining quality. Keep the staining dishes covered with the plastic cover that comes with the staining dish.
- Wash the grid with ddH2O as above.
- Transfer the stained grid to the grid box for further imaging.
- Figure 3C shows an image of wildtype muscle visualized using TEM at 10,000x. In 6-day old flies, VCP RH and AE expressing flies display disrupted actin and mitochondrial structure as compared with wildtype and VCP WT flies (Zhang et al., 2017).
- Select well-aligned samples from the Toluidine Blue staining to be used in thin sectioning (80-90 nm).
- Mitochondrial cross-section size quantification
- Import an ultrastructural electron microscopy image (10,000x magnification) into ImageJ Software (National Institute of Health), Figure 3D.
- After setting the scale (analyze>set scale: 119 pixels = 1 µm), using Polygon selection (Yellow line) trace out each mitochondrion on the image and measure its area (analyze > measure), Figure 3D. All the mitochondria on the image are individually measured.
- At least three images are analyzed for each thorax and 3 thoraxes of each genotype were examined.
- An independent Student’s t-test is used to test for statistical significance between different genotypes.
- In vivo VCP inhibitor treatment
- Dissolve powdered forms of NMS-873 (19.2 mM, stock concentration) and ML240 (25.2 mM, stock concentration) in DMSO as stocks.
- Dilute Stock solution in ethanol/ddH2O (100 µl Ethanol + 600 µl ddH2O) to the desired concentration. Ethanol can help keep the compound in solution. Heat Drosophila food in a microwave for 20 s until it fully melts, and then let it cool down to < 50 °C. Add the chemical in DMSO/ethanol H2O to the food along with 10 µl of food dye per 4 ml of fly food, and hand-mix it until the color appears homogenous. Vials with cotton plugs can be dried at room temperature if food seems wet. DMSO/ethanol in comparable amounts is used as a vehicle control.
- Put Drosophila parents of desired genotypes in food containing DMSO/ethanol or inhibitors for 3 days to lay eggs and then remove them. Progeny growth then occurs in the presence of vehicle control or test compound.
- Immediately after eclosion of adults (hatch from their pupal case), transfer these adults to freshly prepared food containing the same concentration of DMSO or inhibitor, and feed the flies for the desired number of days before assaying them.
- VCP inhibitors feeding significantly reversed the muscle disintegration, muscle cell death and ultrastructural mitochondrial defects in VCP RH and AE flies (Zhang et al., 2017).
Recipes
- Muscle Dissection Fixative Buffer (500 µl)
100 µl 20% paraformaldehyde
400 µl Schneider’s Buffer
Store at 4 °C
Note: Preferably prepare fresh each experiment. - Dissection Buffer
0.1% Triton X-100
1x PBS Buffer
Store at room temperature - TUNEL Blocking Buffer
50 mM Tris-Cl (pH = 7.4)
188 mM NaCl
0.2% Triton X-100
1% BSA
Store at 4 °C - 0.2 M Phosphate Buffer (pH = 7.4)
Solution X: 3.516 g Na2HPO4·2H2O/100 ml ddH2O
Solution Y: 2.76 g NaH2PO4·H2O/100 ml ddH2O
Mix 40.5 ml of Solution X with 9.5 ml of Solution Y, then get 50 ml of 0.2 M Phosphate Buffer. The pH should be 7.4
Store at room temperature - 10 ml EM Fixative Buffer
1.25 ml 8% glutaraldehyde
0.625 ml 16% paraformaldehyde
5.0 ml 0.2 M phosphate buffer
3.125 ml ddH2O Sterile
Store at 4 °C - Epon Mix (Medium)
Embed812 20 ml
DDSA (dodecenyl succinic anhydride) 16 ml
NMA (nadic methyl anhydrate) 8 ml
BDMA (benzyldimethylamine) 0.9 ml (Add fresh for each embedding)
Mix thoroughly at least 1 h before embedding
Can be stored at room temperature in light-proof container. Preferably prepare fresh each time - 1% Toluidine Blue Staining Solution
1 g Toluidine blue powder
1 g Sodium Borate
100 ml of ddH2O
0.22 µm filtered before use.
Store at room temperature in light-proof container - Uranium Acetate Staining Solution
4 g Uranyl acetate
100 ml ddH2O
Dissolve fully (heat up to 70 °C to facilitate dissolving)
0.22 µm filtered before use
Store at room temperature in light-proof acrylic storage container that protects from beta radiation - Lead Citrate Staining Solution
Boil 100 ml ddH2O to get rid of CO2 in a small beaker covered with aluminum foil let it cool to room temperature
1.33 g lead nitrate
1.76 g sodium citrate
30 ml CO2-free ddH2O
8 ml 1 N NaOH (4 g NaOH in 100 ml ddH2O)
Add CO2-free ddH2O to a total volume of 50 ml
Store at 4 °C in light-proof container
Acknowledgments
We are grateful to the generous support from the National Institute of Health (National Institute on Aging), Glenn Foundation for Medical Research, the Natalie R. and Eugene S. Jones Fund in Aging and Neurodegenerative Disease Research, Kenneth Glenn Family Foundation, funds from the UCLA Laurie and Steven Gordon Commitment to Cure Parkinson’s Disease, and Renee and Meyer Luskin Family Fund.
We thank Rosaline Young, Mark Dodson, Hansong Deng, and Jina Yun for developing and optimizing the protocol (Clark et al., 2006; Deng et al., 2008; Yun et al., 2014).
Competing interests
We have no competing interests.
References
- Abramzon, Y., Johnson, J. O., Scholz, S. W., Taylor, J. P., Brunetti, M., Calvo, A., Mandrioli, J., Benatar, M., Mora, G., Restagno, G., Chio, A. and Traynor, B. J. (2012). Valosin-containing protein (VCP) mutations in sporadic amyotrophic lateral sclerosis. Neurobiol Aging 33(9): 2231. e1-2231.e6.
- Bartolome, F., Wu, H. C., Burchell, V. S., Preza, E., Wray, S., Mahoney, C. J., Fox, N. C., Calvo, A., Canosa, A., Moglia, C., Mandrioli, J., Chio, A., Orrell, R. W., Houlden, H., Hardy, J., Abramov, A. Y. and Plun-Favreau, H. (2013). Pathogenic VCP mutations induce mitochondrial uncoupling and reduced ATP levels. Neuron 78(1): 57-64.
- Clark, I. E., Dodson, M. W., Jiang, C., Cao, J. H., Huh, J. R., Seol, J. H., Yoo, S. J., Hay, B. A. and Guo, M. (2006). Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature 441(7097): 1162-1166.
- Chang, Y. C., Hung, W. T., Chang, H. C., Wu, C. L., Chiang, A. S., Jackson, G. R. and Sang, T. K. (2011). Pathogenic VCP/TER94 alleles are dominant actives and contribute to neurodegeneration by altering cellular ATP level in a Drosophila IBMPFD model. PLoS genetics 7(2): e1001288.
- de Bot, S. T., Schelhaas, H. J., Kamsteeg, E. J. and van de Warrenburg, B. P. (2012). Hereditary spastic paraplegia caused by a mutation in the VCP gene. Brain 135(Pt 12): e223.
- Deng, H., Dodson, M. W., Huang, H. and Guo, M. (2008). The Parkinson's disease genes pink1 and parkin promote mitochondrial fission and/or inhibit fusion in Drosophila. Proc Natl Acad Sci U S A 105(38): 14503-14508.
- Gonzalez, M. A., Feely, S. M., Speziani, F., Strickland, A. V., Danzi, M., Bacon, C., Lee, Y., Chou, T. F., Blanton, S. H., Weihl, C. C., Zuchner, S. and Shy, M. E. (2014). A novel mutation in VCP causes Charcot-Marie-Tooth Type 2 disease. Brain 137(Pt 11): 2897-2902.
- Ju, J. S., Fuentealba, R. A., Miller, S. E., Jackson, E., Piwnica-Worms, D., Baloh, R. H. and Weihl, C. C. (2009). Valosin-containing protein (VCP) is required for autophagy and is disrupted in VCP disease. J Cell Biol 187(6): 875-888.
- Kandul, N. P., Zhang, T., Hay, B. A. and Guo, M. (2016). Selective removal of deletion-bearing mitochondrial DNA in heteroplasmic Drosophila. Nat Commun 7: 13100.
- Kim, N. C., Tresse, E., Kolaitis, R. M., Molliex, A., Thomas, R. E., Alami, N. H., Wang, B., Joshi, A., Smith, R. B., Ritson, G. P., Winborn, B. J., Moore, J., Lee, J. Y., Yao, T. P., Pallanck, L., Kundu, M. and Taylor, J. P. (2013). VCP is essential for mitochondrial quality control by PINK1/Parkin and this function is impaired by VCP mutations. Neuron 78(1): 65-80.
- Kimonis, V. E., Fulchiero, E., Vesa, J. and Watts, G. (2008a). VCP disease associated with myopathy, Paget disease of bone and frontotemporal dementia: review of a unique disorder. Biochim Biophys Acta 1782(12): 744-748.
- Kimonis, V. E., Mehta, S. G., Fulchiero, E. C., Thomasova, D., Pasquali, M., Boycott, K., Neilan, E. G., Kartashov, A., Forman, M. S., Tucker, S., Kimonis, K., Mumm, S., Whyte, M. P., Smith, C. D. and Watts, G. D. (2008b). Clinical studies in familial VCP myopathy associated with Paget disease of bone and frontotemporal dementia. Am J Med Genet. Part A 146A (6): 745-757.
- Kimura, Y., Fukushi, J., Hori, S., Matsuda, N., Okatsu, K., Kakiyama, Y., Kawawaki, J., Kakizuka, A. and Tanaka, K. (2013). Different dynamic movements of wild-type and pathogenic VCPs and their cofactors to damaged mitochondria in a Parkin-mediated mitochondrial quality control system. Genes Cells 18(12): 1131-1143.
- Ma, P., Yun, J., Deng, H. and Guo, M. (2018). Atg1 mediated autophagy suppresses tissue degeneration in pink1/parkin mutants by promoting mitochondrial fission in Drosophila. Mol Biol Cell: mbcE18040243.
- Meyer, H., Bug, M. and Bremer, S. (2012). Emerging functions of the VCP/p97 AAA-ATPase in the ubiquitin system. Nat Cell Biol 14(2): 117-123.
- Ritson, G. P., Custer, S. K., Freibaum, B. D., Guinto, J. B., Geffel, D., Moore, J., Tang, W., Winton, M. J., Neumann, M., Trojanowski, J. Q., Lee, V. M., Forman, M. S. and Taylor, J. P. (2010). TDP-43 mediates degeneration in a novel Drosophila model of disease caused by mutations in VCP/p97. J Neurosci 30(22): 7729-7739.
- Ritz, D., Vuk, M., Kirchner, P., Bug, M., Schutz, S., Hayer, A., Bremer, S., Lusk, C., Baloh, R. H., Lee, H., Glatter, T., Gstaiger, M., Aebersold, R., Weihl, C. C. and Meyer, H. (2011). Endolysosomal sorting of ubiquitylated caveolin-1 is regulated by VCP and UBXD1 and impaired by VCP disease mutations. Nat Cell Biol 13(9): 1116-1123.
- Watts, G. D., Wymer, J., Kovach, M. J., Mehta, S. G., Mumm, S., Darvish, D., Pestronk, A., Whyte, M. P. and Kimonis, V. E. (2004). Inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia is caused by mutant valosin-containing protein. Nat Genet 36(4): 377-381.
- Weihl, C. C., Pestronk, A. and Kimonis, V. E. (2009). Valosin-containing protein disease: inclusion body myopathy with Paget's disease of the bone and fronto-temporal dementia. Neuromuscul Disord 19(5): 308-315.
- Yun, J., Cao, J. H., Dodson, M. W., Clark, I. E., Kapahi, P., Chowdhury, R. B. and Guo, M. (2008). Loss-of-function analysis suggests that Omi/HtrA2 is not an essential component of the PINK1/PARKIN pathway in vivo. J Neurosci 28(53): 14500-14510.
- Yun, J., Puri, R., Yang, H., Lizzio, M. A., Wu, C., Sheng, Z. H. and Guo, M. (2014). MUL1 acts in parallel to the PINK1/parkin pathway in regulating mitofusin and compensates for loss of PINK1/parkin. Elife 3: e01958.
- Zhang, T., Mishra, P., Hay, B. A., Chan, D. and Guo, M. (2017). Valosin-containing protein (VCP/p97) inhibitors relieve Mitofusin-dependent mitochondrial defects due to VCP disease mutants. Elife 6: e17834.
Article Information
Copyright
Zhang et al. This article is distributed under the terms of the Creative Commons Attribution License (CC BY 4.0).
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Zhang, T., Hay, B. A. and Guo, M. (2020). Generation, Analyzing and in-vivo Drug Treatment of Drosophila Models with IBMPFD. Bio-protocol 10(10): e3621. DOI: 10.21769/BioProtoc.3621.
- Zhang, T., Mishra, P., Hay, B. A., Chan, D. and Guo, M. (2017). Valosin-containing protein (VCP/p97) inhibitors relieve Mitofusin-dependent mitochondrial defects due to VCP disease mutants. Elife 6: e17834.
Category
Neuroscience > Nervous system disorders > Animal model
Molecular Biology > Protein > Protein-protein interaction
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









