- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Quantification of Fatty Acids in Mammalian Tissues by Gas Chromatography–Hydrogen Flame Ionization Detection
Published: Vol 10, Iss 9, May 5, 2020 DOI: 10.21769/BioProtoc.3613 Views: 6234
Reviewed by: Khyati Hitesh ShahXiaofei LiangAksiniya AsenovaAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Lipidomics Workflow for Analyzing Lipid Profiles Using Multiple Reaction Monitoring (MRM) in Liver Homogenate of Mice with Non-alcoholic Steatohepatitis (NASH)
Hai Ning Wee [...] Jianhong Ching
Jul 5, 2023 2448 Views

Computational Analysis of Plasma Lipidomics from Mice Fed Standard Chow and Ketogenic Diet
Amy L. Seufert [...] Brooke A. Napier
Sep 20, 2023 2607 Views

Quantitative Determination of Cholesterol Hydroxylase Specificities by GC–MS/MS in Living Mammalian Cells
Hodaka Saito [...] Yoshio Yamauchi
Jan 20, 2024 2369 Views
Abstract
In mammalian organisms, fatty acids (FAs) exist mostly in esterified forms, as building blocks of phospholipids, triglycerides, and cholesteryl esters, while some exist as non-esterified free FAs. The absolute quantification of FA species in total lipids or in a specific lipid class is critical in lipid-metabolism studies. To quantify FAs in biological samples, gas chromatography–hydrogen flame ionization detection (GC-FID)-based methods have been used as highly robust and reliable techniques. Prior to GC-FID analysis, FAs need to be derivatized to volatile FA methyl esters (FAMEs). The derivatization of unsaturated FAs using classical derivatization methods that rely on high reaction temperature requires skill; consequently, the quantification results are often unreliable. The recently available FA-methylation procedure rapidly and reliably derivatizes a variety of FA species, including poly-unsaturated FAs (PUFAs). To analyze FAs in mammalian tissue samples, lipid extraction and fractionation are also critical for robust analysis. In this report, we describe a whole protocol for the GC-FID-based FA quantification of mammalian tissue samples, including lipid extraction, fractionation, derivatization, and quantification. The protocol is useful when various FAs, especially unsaturated FAs, need to be reliably quantified.
Keywords: Fatty acidsBackground
A fatty acid (FA) is a carboxylic acid with an aliphatic chain, and FAs are classified according to their chain lengths (short, medium, long, very long) and the number of intramolecular double bonds (saturated, mono- or poly-unsaturated). In mammalian organisms, FAs exist mostly in esterified forms, such as phospholipids (PLs), triglycerides (TGs), and cholesteryl esters. PLs are major components of biological membranes, while TGs are important as concentrated energy stores, and cholesteryl esters play roles in cholesterol metabolism (Hishikawa et al., 2014; Nielsen et al., 2014; Hui and Howles, 2005; Van Meer et al., 2008).
Mammalian PLs are classified into several major classes, including phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylserine (PS), phosphatidylinositol (PI), phosphatidic acid (PA), cardiolipin (CL), and sphingomyelin (SM), each of which contain a number of different molecular species due to the diversity of the two fatty acyl groups (i.e., FAs) at their sn-1 and sn-2 positions (except for CL and SM, which contain four and one fatty acyls, respectively). PLs are known to have asymmetric FA profiles between the sn-1 and sn-2 positions; sn-1 prefers saturated or mono-unsaturated FAs, while sn-2 can be substituted by a variety of FAs, including poly-unsaturated FAs. The molecular basis and biological significance of the diversity in the FA compositions of membrane PLs had not been elucidated over many years, but recent studies have demonstrated critical roles for several lysophospholipid acyltransferases (LPLATs), which are enzymes that incorporate distinct FAs into phospholipids (Hashidate-Yoshida et al., 2015; Shindou et al., 2017; Iizuka-Hishikawa et al., 2017). In these reports, phenotypic analysis of gene-targeted (knock-out) mice for the LPLAT gene(s) was performed along with the lipidomic profiling of various tissues. To this end, two methodologies, namely liquid-chromatography–mass-spectrometry-based (LC-MS-based) PL molecular-species profiling (comparative/differential analysis) and gas chromatography–hydrogen flame ionization detection (GC-FID) for the absolute quantification of FA compositions, have been adopted. These two methods are complementary; while the LC-MS method tracks relative PL changes at the molecular-species level, the GC-FID method provides absolute comparisons of various FAs in total lipids, PLs, and neutral lipids.
GC-FID is one of the most robust techniques for detecting various fatty acids. Since GC can only analyze volatile compounds, FAs require prior derivatization, and are typically methylated to volatile FA methyl esters (FAMEs). Classical methylation protocols rely on high-temperature derivatization reactions (Vorbeck et al., 1961; Liu, 1994), which require time and skill. An alternative derivatization procedure, which can be performed rapidly with mild heating has been established (Ichihara et al., 1996; Ichihara and Fukubayashi, 2010), and is now commercially available in the form of a kit. As a GC detector, FID is favorably used for the robust detection of FAMEs. FID, which shows proportional responses to compound quantities with broad linear dynamic ranges, is suited to the sensitive detection of various organic compounds including FAMEs. MS-based detection (e.g., GC-MS) is a more-sensitive alternative to FID, but FID is advantageous due to its stability, dynamic range, and its introduction and maintenance costs.
In this report, we describe a whole protocol for quantifying the FAs of murine tissues by GC-FID. The protocol describes the determination of total FAs, which is the sum of the esterified FAs and non-esterified free FAs (FFAs) in the total lipids, and the esterified fatty acids in neutral or phospholipid fractions after solid-phase extraction (Egberts and Buiskool, 1988).
Materials and Reagents
- Pipette tips: 200, 1,200, and 5,000 µl (GILSON, catalog numbers: F167103 , F171600 , F161571 )
- 10-µl pipette tips (GP-LTS-A-10 µL-960/10; RAININ, catalog number: 30389270 )
- 0.5-ml microtubes (BIO-BIK, catalog number: 103001 )
- 2-ml pulverizing tubes with stainless steel inner lids (Bio Medical Science, catalog number: MT020-01HS )
- Borosilicate glass test tubes: 16 x 125 mm and 13 x 100 mm (IWAKI, catalog numbers: 9835-1612 , 9832-1310 )
- Test tube caps: 13 mm and 16 mm (Fisher Scientific, catalog numbers: 14-376-69 , 14-376-83 )
- 50-µl GC vial inserts with bottom spring (Shimadzu GLC, catalog number: 98058 )
- 2.0-ml autosampler vials (Chromacol, catalog number: 2-SV )
- Septa for 2.0-ml vials (Shimadzu, catalog number: 221-41239-91 )
- Screw caps for 2.0-ml vials (Shimadzu, catalog number: 228-15653-91 )
- Screw caps with closed tops for 2.0-ml vials (Chromacol, catalog number: 8-SCS ) (for storage)
- Mouse tissue
- InertSep NH2-cartridge: 200 mg/3 ml (GL Science, catalog number: 5010-61602 )
- Supelco 37-component FAME mix (Sigma-Aldrich, catalog number: CRM47885 )
- cis-4,10,13,16-Docosatetraenoic acid methyl ester [DTA (C22:4n-6) FAME] (Cayman, catalog number: 10006866 )
- cis-7,10,13,16,19-Docosapentaenoic methyl ester [DPA (C22:5n-3) FAME] (Sigma-Aldrich, catalog number: 47563-U )
- 4Z,7Z,10Z,13Z,16Z-Docosapentaenoic methyl ester [DPA (C22:5n-6) FAME] (Nu-Chek Prep, Inc., catalog number: U-102-M )
- Tricosanoic acid (C23:0 FA) (Sigma-Aldrich, catalog number: T6543 )
- Methanol (for high performance liquid chromatography) (FUJIFILM Wako, catalog number: 132-06471 )
- Chloroform (Guaranteed Reagent) (FUJIFILM Wako, catalog number: 038-02606 )
- Dichloromethane (Guaranteed Reagent) (FUJIFILM Wako, catalog number: 135-02441 )
- Acetone (Guaranteed Reagent) (FUJIFILM Wako, catalog number: 016-00346 ) (for GC syringe washing)
- Hexane (Guaranteed Reagent) (FUJIFILM Wako, catalog number: 085-00416 )
- 2-Propanol (Guaranteed Reagent) (FUJIFILM Wako, catalog number: 166-04836 )
- Acetic acid (Guaranteed Reagent) (FUJIFILM Wako, catalog number: 017-00256 )
- Diethyl ether (Guaranteed Reagent) (FUJIFILM Wako, catalog number: 055-01155 )
- 28% Ammonia solution (FUJIFILM Wako, catalog number: 016-03146 )
- Dulbecco’s phosphate buffered saline (PBS) (10x) (Sigma-Aldrich, catalog number: D1408 )
- Ultra-pure water (MilliQ)
- He (> 99.999 vol%)
- H2 (> 99.999 vol%)
- Liquid nitrogen
- Fatty acid methylation kit (Nacalai Tesque, catalog number: 16962-04 )
Methylation Reagent A
Methylation Reagent B
Methylation Reagent C
Isolation Reagent - Fatty acid methyl ester purification kit (Nacalai Tesque, catalog number: 16961-14 )
Conditioning solution
Washing solution
Eluting solution
SPE cartridge column - Chloroform:2-propanol (2:1, v:v) (see Recipes)
- 2% acetic acid in diethyl ether (see Recipes)
- 2.8% ammonia in methanol (see Recipes)
- PBS (see Recipes)
- C23:0 FA (5 mg/ml stock and 1 mg/ml working solutions) (see Recipes)
- Standard FAME mix (serial dilution) (see Recipes)
- DTA (C22:4n-6) FAME (~1 mg/ml stock solution) (see Recipes)
- DPA (C22:5n-6) FAME (~10 mg/ml and ~1 mg/ml stock solutions) (see Recipes)
- DPA (C22:5n-3) FAME (~1 mg/ml stock solution) (see Recipes)
- DTA and DPA FAME mix (see Recipes)
Equipment
- Pipetman (Gilson, models: P5000, P1000 and P200)
- Glass graduated cylinder
- Pipet-Lite LTS Pipette L-10XLS+ (RAININ, catalog number: 17014388 )
- Forceps
- Vortex mixer (Scientific Industries, catalog number: G-560 )
- Vacuum evaporator (Sakuma, catalog number: EC-95C3T )
- Water bath (TAITEC, model: Temperature Chamber Thermo Minder SD ) (for 37 °C incubation)
- Centrifuge with swing rotor (Hitachi, model: Himac CF8DL )
- CAPSULEFUGE microcentrifuge (TOMY, catalog number: PMC-060 )
- Gas chromatography (GC) system equipped with a flame ionization detector (FID) (Shimadzu, model: GC2010 Plus )
- FAMEWAX GC capillary column, 30 m x 0.25 mm; ID, 0.25 µm (Restek, catalog number: 12497 )
- Automatic cryogenic pulverizer (Tokken, Inc., model: AUTOMILL TK-AM5 )
- Stainless-steel crusher (Tokken, Inc., catalog number: SK-100-DLC10 )
- Rotator (TAITEC, model: RT-50 )
Software
- GC Solution Software, version 2.44 (Shimadzu)
- Microsoft Excel
Procedure
The entire experimental workflow described in this protocol is shown in Figure 1. The blue arrow is followed when determining only total FAs, while the green arrows are followed when determining FAs in neutral-lipid and/or phospholipid fractions.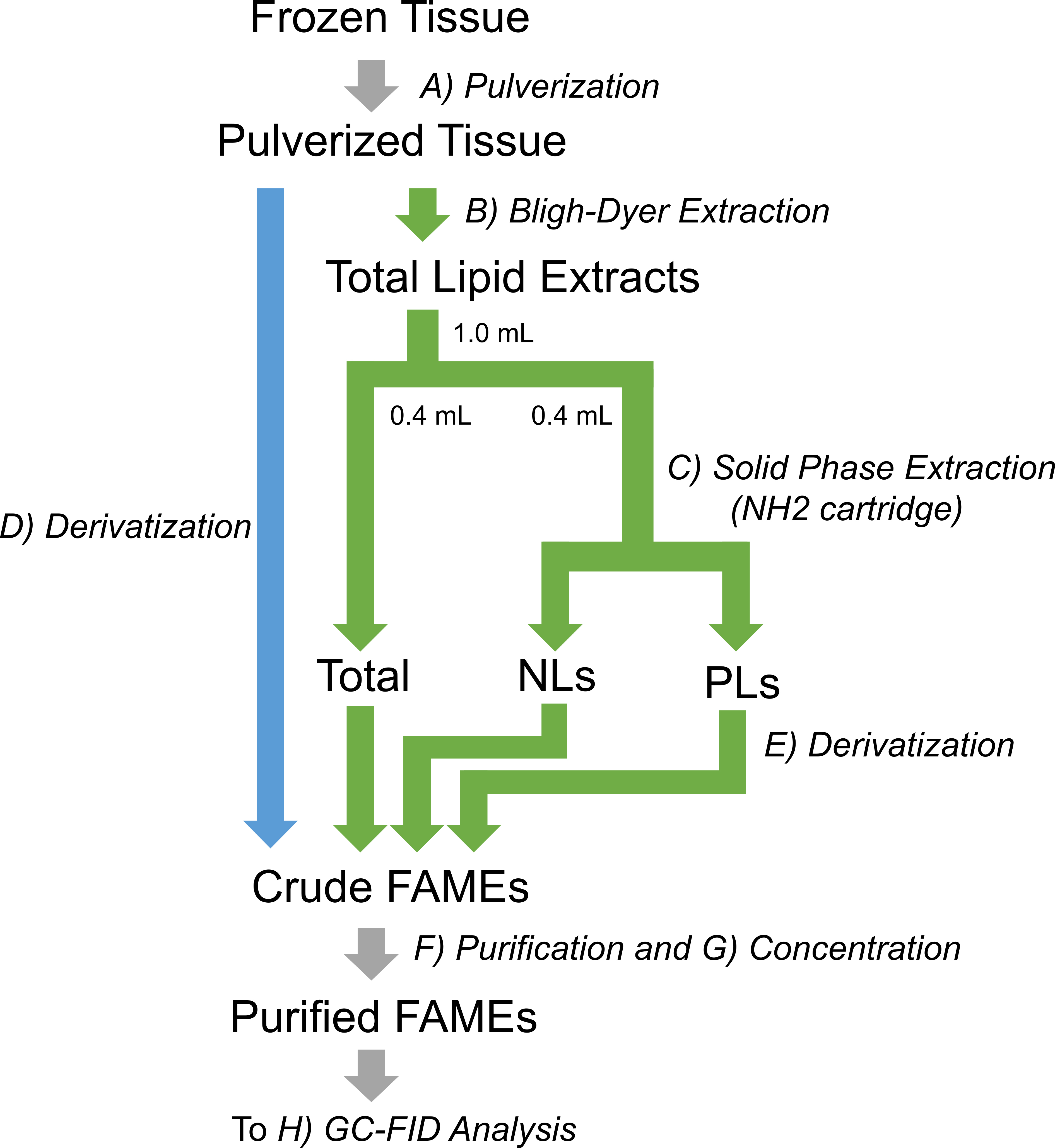
Figure 1. Workflow for the quantification of FAs from tissue samples by GC-FID. Frozen tissues are cryogenically pulverized. The pulverized tissue is directly subjected to the derivatization procedure (blue path) when only total FA quantification is required. To quantify FAs in neutral-lipid (NL) and phospholipid (PL) fractions, as well as total lipids (total), the total lipids are extracted from the pulverized tissue using the Bligh-Dyer method, fractionated by solid-phase extraction, and then derivatized (green route).
- Tissue pulverization
- Place frozen mouse tissue (~50 mg or less) and a stainless-steel crusher into a 2.0-ml pulverizing tube with a stainless steel inner lid, and cool in liquid nitrogen.
- Shake the tubes using an automatic cryogenic pulverizer (900-1,500 rpm, 20 s). Repeat several times until fully pulverized.
- If only total FA analysis is required, continue to Procedure D. When phospholipid and neutral lipid fractions also need to be analyzed, continue to Procedure B.
- Bligh-Dyer total lipid extraction from pulverized tissue (Bligh and Dyer, 1959)
- Add 800 µl of methanol to the tube containing the pulverized tissue and the crusher.
- Gently rotate at 4 °C for 1 h.
- Remove crusher from the tube with forceps.
- Decant the tube contents into a glass test tube (16 x 125 mm) containing 1 ml of PBS.
- To recover all tissue, wash the pulverizing tube serially with 1.0 ml of methanol, 1.25 ml of chloroform, and 0.7 ml of methanol into the glass test tube.
Note: The glass test tube will contain pulverized tissue, 2.5 ml of methanol, 1.25 ml of chloroform, and 1 ml of PBS. - Vortex for 2 min at maximum speed.
- Settle the glass test tube at room temperature for 10 min.
- Add 1.25 ml of chloroform to the glass test tube and vortex for 2 min.
- Add 1.25 ml of PBS to the glass test tube and vortex for 2 min.
- Centrifuge the glass test tube at 1,000 x g at room temperature using the swing rotor for 10 min.
- Transfer the lower (organic) phase to a new glass test tube (16 x 125 mm).
- For full lipid recovery, add 1.0 ml of chloroform to the remaining aqueous phase, vortex for 2 min, centrifuge for 10 min, collect the lower phase, and combine it with the first extract.
- Evaporate the glass test tube containing the extracts to dryness using a vacuum evaporator. This step typically takes ~1 h for ~50 mg of tissue from Procedure A, although this depends on the tissue type and evaporator performance.
- Add 1 ml of chloroform and vortex until the extracts are dissolved.
- Continue to Procedure C.
- Preparing neutral-lipid and phospholipid fractions by solid phase extraction
- Remove a 0.4-ml aliquot from the 1-ml total lipid extract (chloroform solution) prepared in Procedure B and transfer it to a new glass test tube (13 x 100 mm) labeled ‘Total Lipids’. For quantitative accuracy, the pipette should be pre-wetted with chloroform.
- Pre-equilibrate an InertSep NH2 cartridge (200 mg/3 ml) with 9 ml (3 x 3 ml) of hexane.
- Place a new glass test tube (16 x 125 mm) labeled ‘Neutral Lipids’ at the bottom of cartridge.
- Using a pre-wetted pipette, load 0.4 ml from the remaining volume (i.e., ~0.6 ml) of the total lipid extract prepared in Procedure B onto the NH2 cartridge. Collect the flow-through fraction in the ‘Neutral Lipids’ test tube.
- Elute with 9 ml (3 x 3 ml) of chloroform:2-propanol (2:1) into the ‘Neutral Lipids’ test tube.
- Remove the ‘Neutral Lipids’ test tube from the cartridge.
- Wash the cartridge with 9 ml (3 x 3 ml) of 2% acetic acid in diethyl ether to remove the FFA.
Note: Since the eluent often contains interfering materials of plasticware origin that hamper C16:0 and C18:0 FA analyses, we do not recommend using this fraction for FFA determination. - Place a new glass test tube (16 x 125 mm) labeled ‘Phospholipids’ at the bottom of the cartridge.
- Elute with 9 ml (3 x 3 ml) of 2.8% ammonia in methanol into the glass test tube.
- Remove the glass test tube containing the phospholipids from the cartridge.
- Retain the ‘Total Lipids’, ‘Neutral Lipids’, and ‘Phospholipids’ test tubes for derivatization.
- Continue to Procedure E.
- Extracting and derivatizing total FA from pulverized tissue samples
- Add 500 µl of Methylation Reagent A to the pulverization tube containing freshly pulverized tissue and a crusher.
- Add 10 µl of a 1-mg/ml C23:0 FA stock solution, using a pre-wetted pipette.
Note: This step is critical for quantitative accuracy. Pre-wetting should be carried out until no further liquid drips are observed from the pipette tip. - Mix using the rotator for 1 h at 3-6 rpm and 4 °C.
- Remove the crusher from the tube with forceps.
- Decant the contents of the tube into a glass test tube (13 x 100 mm).
- Add 500 µl of Methylation Reagent B and vortex for 5 s.
Note: Use test tube caps for vortexing and incubation. - Incubate for 1 h in a water bath at 37 °C.
- Add 500 µl of Methylation Reagent C and vortex for 5 s.
- Incubate for 20 min in a water bath at 37 °C.
- Add 1 ml of the Isolation Reagent and vortex for 20 s.
- Wait until phase separation is complete and then transfer the upper phase to a new glass test tube (13 x 100 mm).
- Add 1 ml of water to the glass test tube and vortex for 20 s.
- Wait until phase separation is complete and then transfer the upper phase to a new glass test tube (13 x 100 mm).
- Continue to Procedure F.
- Derivatizing lipids dissolved in organic solvents
- Evaporate the samples to be derivatized (i.e., total lipids, neutral lipids, and phospholipids, prepared in Procedure C) to dryness using a vacuum evaporator.
- Add 500 µl of Methylation Reagent A.
- Add 10 µl of a 1-mg/ml C23:0 FA stock solution, using a pre-wetted pipette.
Note: This step is critical for quantitative accuracy. Pre-wetting should be carried out until no further liquid drips are observed from the pipette tip. - Vortex for 20 s.
Note: Use test tube caps for vortexing and incubation. - Add 500 µl of Methylation Reagent B and vortex for 5 s.
- Incubate for 1 h in a water bath at 37 °C.
- Add 500 µl of Methylation Reagent C and vortex for 5 s.
- Incubate for 20 min in a water bath at 37 °C.
- Add 1 ml of the Isolation Reagent and vortex for 20 s.
- Wait until phase separation is complete and then transfer the upper phase to a new glass test tube (13 x 100 mm).
- Add 1 ml of water to the tube and vortex for 20 s.
- Wait until phase separation is complete and then transfer the upper phase to a new glass test tube (13 x 100 mm).
- Continue to Procedure F.
- Purifying FAMEs
- Precondition the SPE cartridge column by passing 3 ml of the conditioning solution through it.
- Load the FAME sample prepared in Procedure D or E (~1 ml) onto the cartridge.
- Wash with 3 mL of washing solution.
- Place a glass test tube (13 x 100 mm) at the bottom of cartridge.
- Elute with 3 mL of the eluting solution.
- If a low FAME concentration is expected then continue to Procedure G. Otherwise continue to Procedure H.
- Concentrating FAMEs
- Evaporate the FAME solution prepared in Procedure F using a vacuum evaporator. This step will take 10-15 min. Extended evaporation after dryness will lose shorter-chain FAMEs and therefore should be avoided; this is significant for FAMEs shorter than C16.
- Reconstitute the sample in a small volume (e.g., 20 µl) of dichloromethane and transfer it to the GC vial insert. The volume should be determined so that the GC-FID signals are within the linear range. Since quantification does not depend on the reconstituted volume, the volume can be varied by sample.
- GC-FID analysis
- Set the column oven, injector, and detector temperatures to 140 °C, 240 °C, and 250 °C, respectively.
- Set the helium carrier gas to a linear velocity of 45 cm/s.
- Analyze the standards (std1 to std4, and the DTA/DPA FAME mix, Recipes 6-10) by injecting a 2-µl volume at a split ratio of 1:25 onto the FAMEWAX column. The oven-temperature program summarized in Table 1 will give good separation (Figure 2).
Note: The cis and trans FAME isomers are combined throughout data processing for C18:1n-9 and C18:2n-6 as they are not resolved in this protocol. This is practical for mammalian tissue samples that contain only cis FA isomers. - Analyze unknown samples with the same GC method (Figure 3). The split ratio and injection volume can be adjusted according to the sample concentration.
Table 1. GC column oven temperature program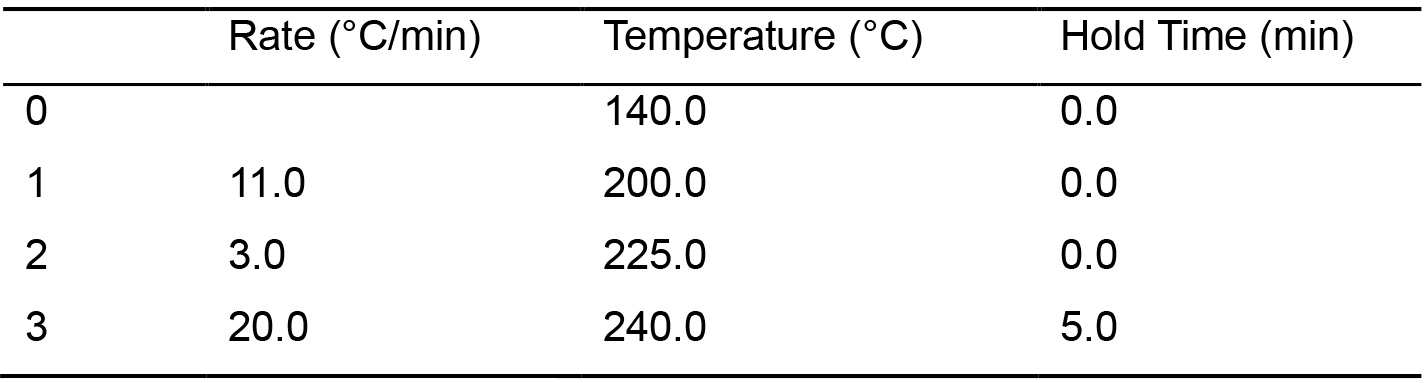
Total program time: 19.54 min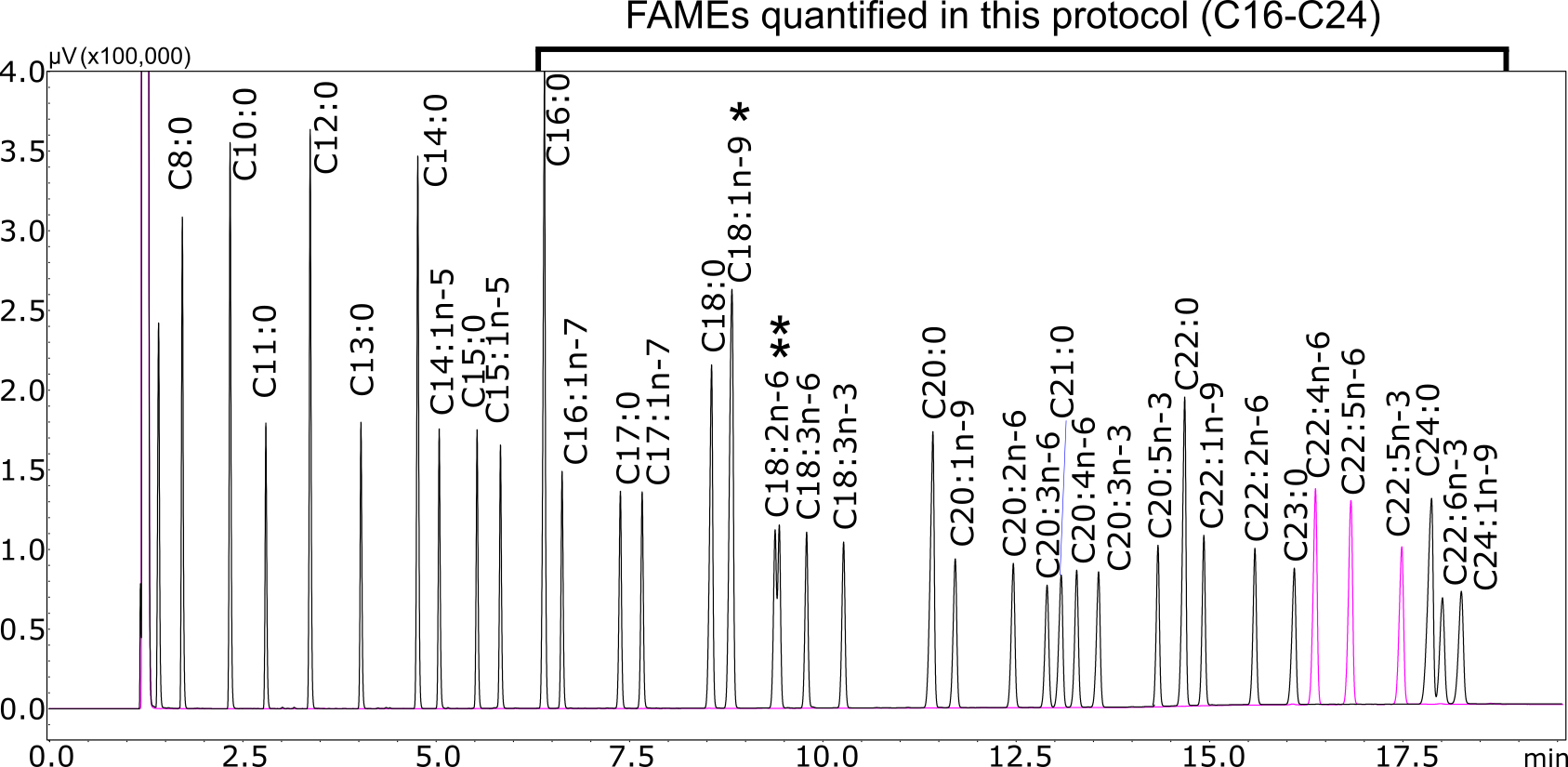
Figure 2. GC-FID traces for FAME standards. Overlaid chromatograms for the Supelco 37-component FAME mix (black) and the DTA/DPA FAME mix (pink) are shown. FAMEs longer than C16 can be quantified using this protocol, as the evaporation step (Procedure G) compromises the quantification of shorter-chain FAMEs. *C18:1n-9 and **C18:2n-6 FAMEs are integrated and quantified as sums of cis and trans isomers, as they are not fully separated.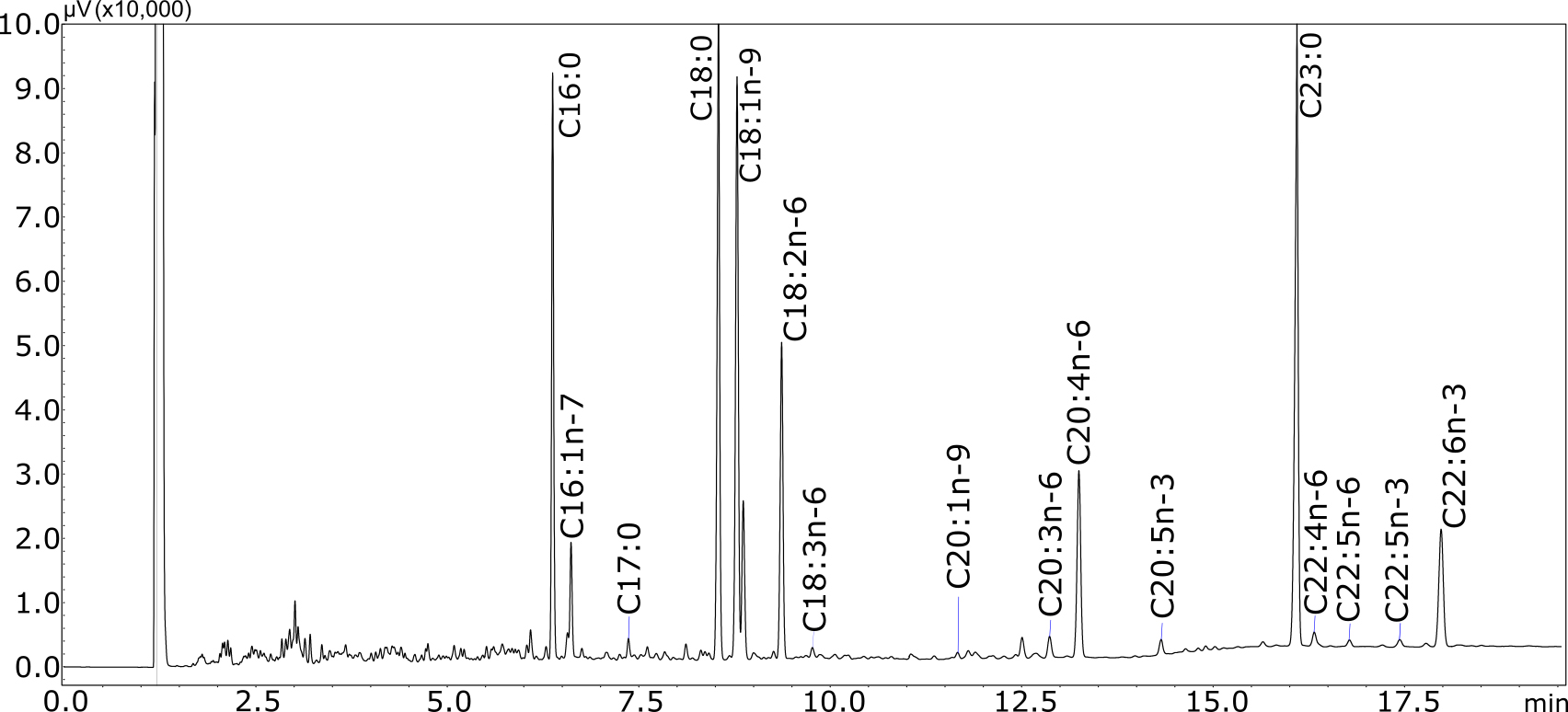
Figure 3. Typical chromatogram for mouse tissue sample. Total lipids were extracted from ~5 mg of E18.5 mouse small intestine by the Bligh-Dyer method, 40% of which were derivatized to give FAMEs. After reconstituting in 20 µl of dichloromethane, 2 µl was injected onto the GC column at a split ratio of 1:25.
Data analysis
- In the Shimadzu GC Solution software, prepare a compound table for quantification using a linear calibration curve (Curve Fit Type: ‘Linear’), by filling the retention times and calibration levels of the 27 target FAMEs (24 FAMEs are from the Supelco 37-component FAME mix, and three additional FAMEs are from the DTA/DPA FAME mix) (Figure 4A, red box). Retention times can be filled according to the peak results obtained during standard analyses. Calibration levels (concentrations) can be set as shown in Table 2.
Note: Dummy (non-zero) calibration levels are set for the three additional FAMEs in the DTA/DPA FAME mix. Once set up, the compound table can be re-used as a template for future analyses, with minor retention-time modification.
Table 2. Concentrations of serially diluted standard FAMEs. Concentrations of *C18:1n-9 and **C18:2n-6 FAMEs are expressed as sums of cis and trans isomers, as these isomers are not separated by GC (see Figure 2).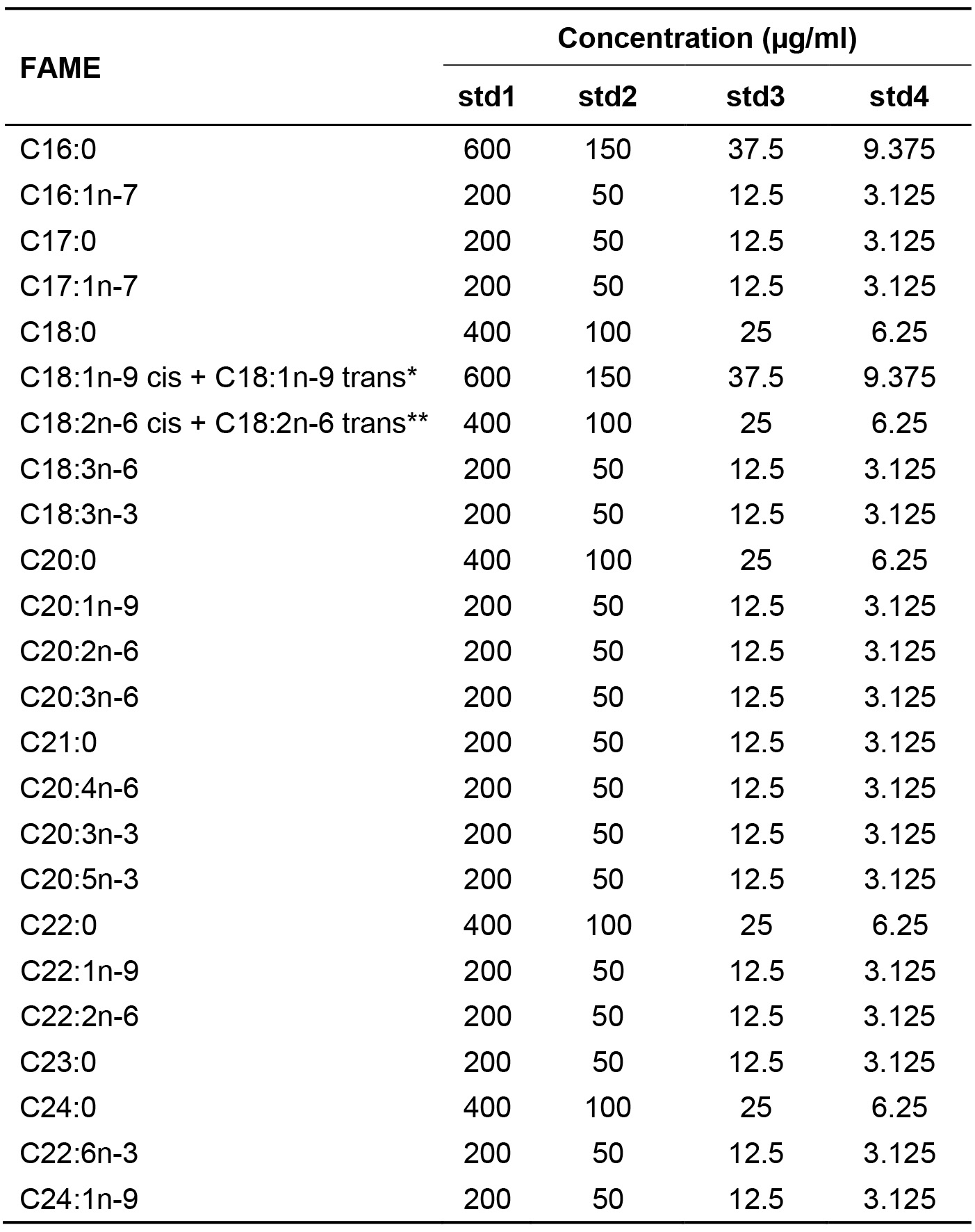
- Process the std1 to std4 data files to generate linear calibration curves with the ‘force origin’ option, which forces the Y-intercept to be zero. Check that linear responses are observed for the 24 FAMEs (Figure 4A, blue box).
- Change ‘Curve Fit Type’ from ‘Linear’ to ‘Manual RF (Linear)’ in the GC Solution software (Figure 4B). This allows the user to modify slope (response factor, RF) values manually. Copy the RF value of C23:0 FAME to those for the DTA/DPA FAMEs to calibrate them as C23:0 FAME-equivalents (Figure 4B, arrow). This approximation is practical and valid due to the characteristics of the FID. As the RF values of all FAMEs are similar, the RF values of other FAMEs can be used as valid approximations.
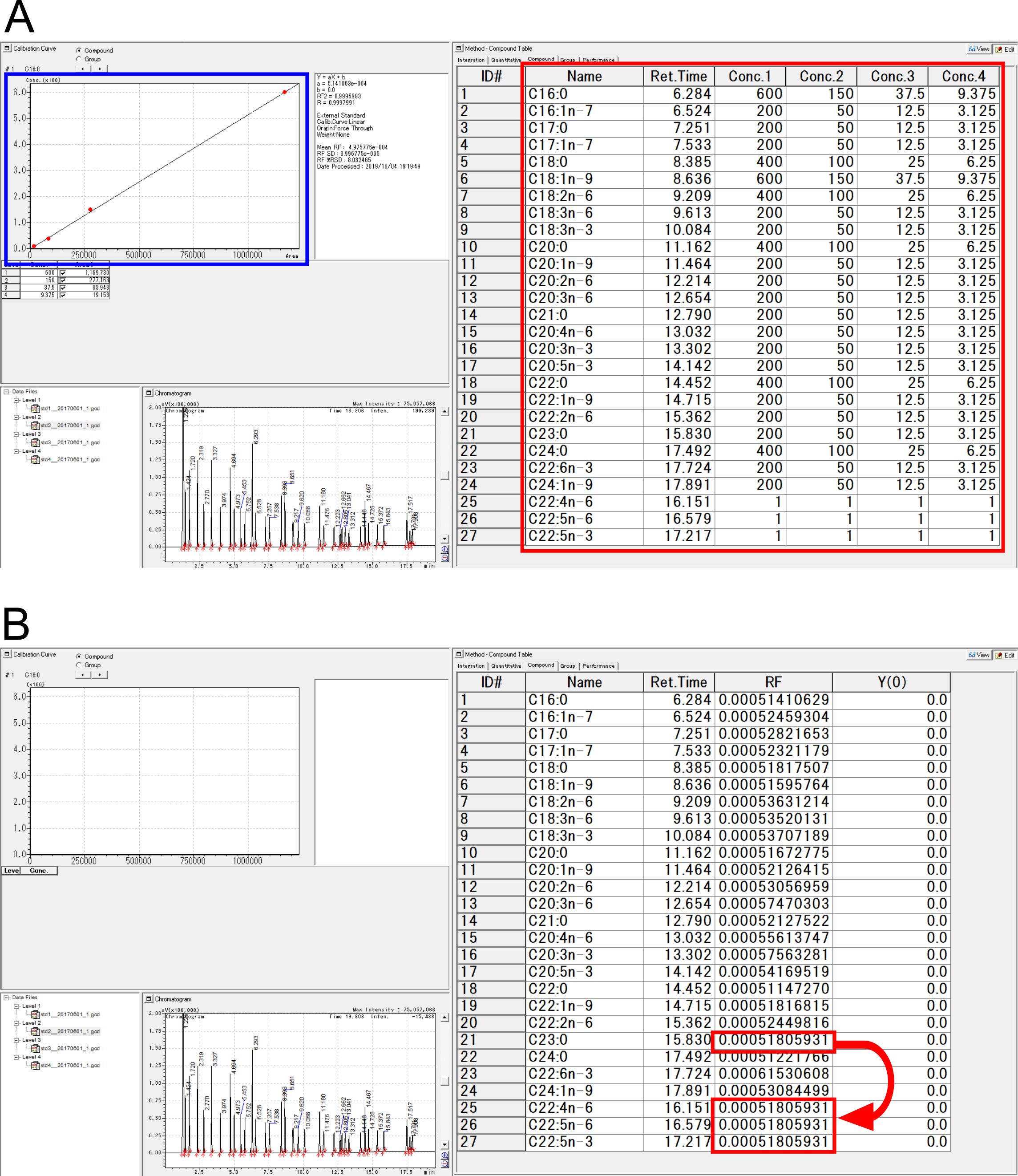
Figure 4. Data processing with the Shimadzu GC Solution software. A. Compound table (red box) and calibration curve (blue box). Calibration curves can be reviewed for linearity after processing the standard samples. B. Calibrating the DTA and DPA FAMEs using the C23:0 FAME. After changing the curve-fit type to ‘Manual RF (Linear)’, the response factor (RF) for C23:0 FAME can be copied to those of the DTA and DPA FAMEs. - Process data files for unknown samples using the prepared calibration settings.
- Check that all peaks are correctly detected; if not, correct either manually or by modifying the peak-detection parameters.
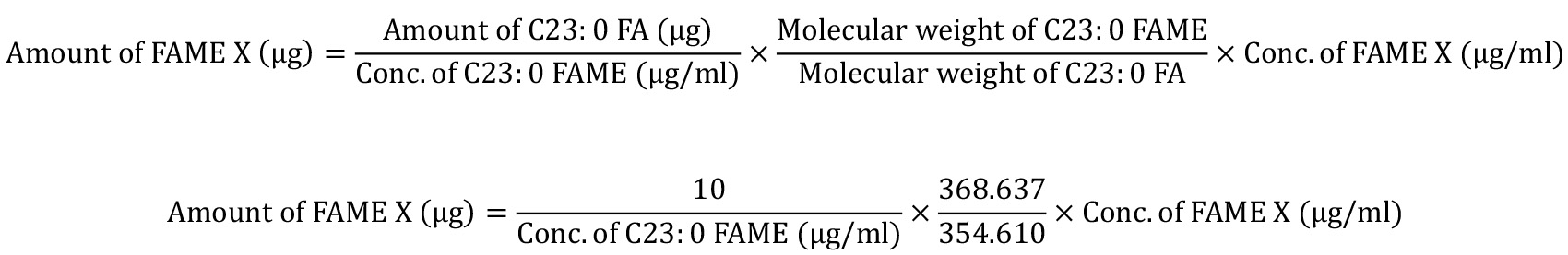
- Copy-and-paste the raw quantification data from GC Solution into an MS Excel worksheet and calculate the absolute amounts of FA in the original sample (Table 3). As the raw quantification data are the FAME concentrations in the GC vials, the absolute amounts of the FAMEs in the sample can be obtained by proportional calculations, using the known amount (i.e., 10 µg) of the C23:0 FA spiked into the samples:
The FAME amounts can then be converted into FA amounts using the molecular weights of the FAMEs and the corresponding FAs (see Supplemental):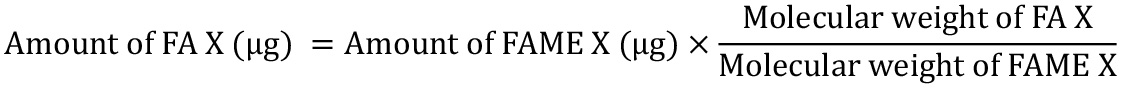
Table 3. Example showing how FA amounts are calculated from raw quantification data. This table uses the quantification results for E18.5 mouse small intestine as an example. Raw quantification data from the GC Solution software, showing FAME concentrations in the GC vial (µg/ml), can be converted into FAME amounts (µg) by proportional calculations, which adjust the amount of C23:0 FAME in the sample to 10.396 µg (equivalent to 10 µg of C23:0 FA). FAME amounts can be further converted into FA amounts using molecular-weight ratios. Values shown in red are constants that are not affected by the various samples. When a part of a sample is subjected to FAME derivatization, further proportional calculations are necessary to determine the amounts of FA in the initial sample.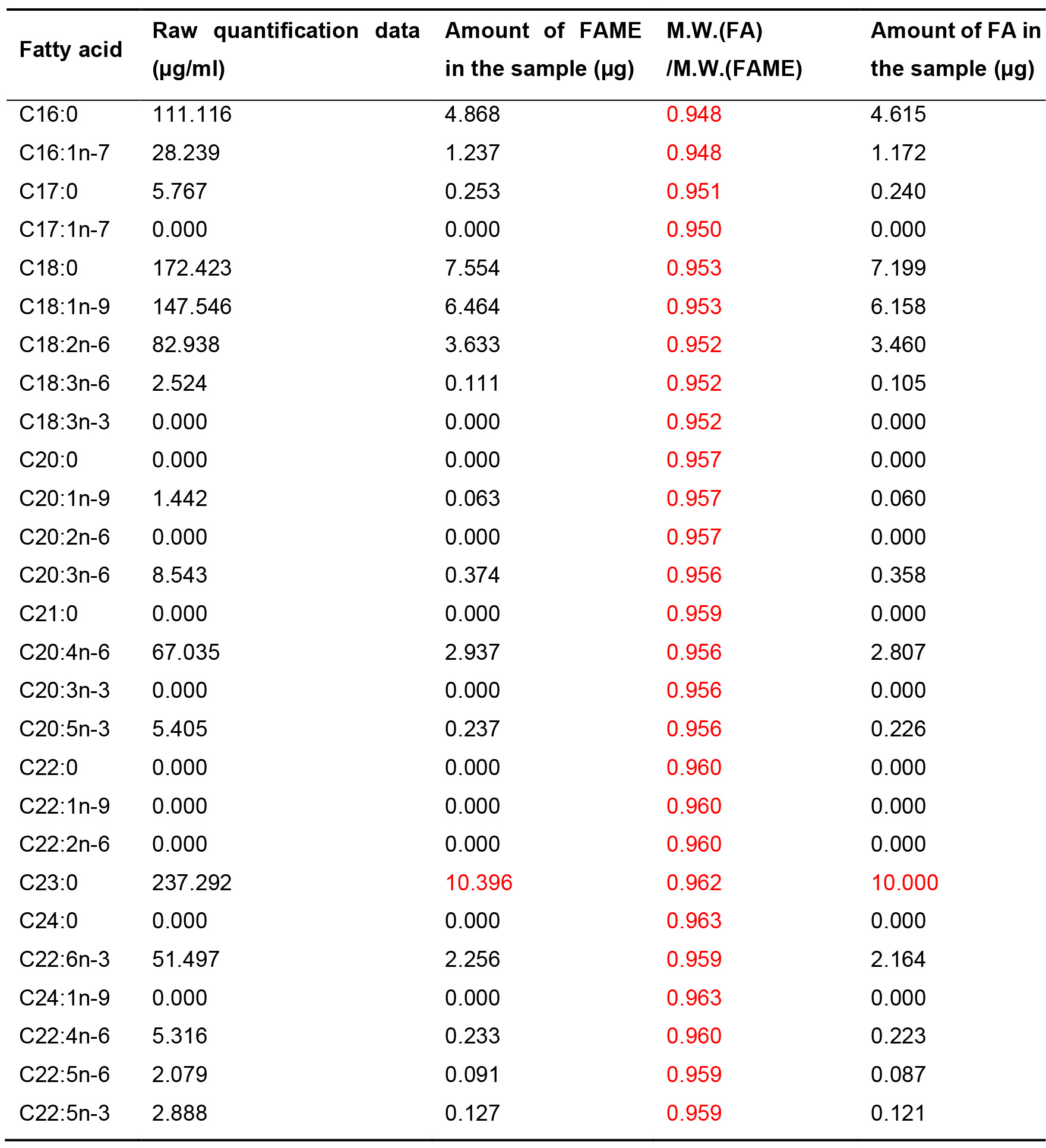
- Perform proportional calculations to determine the amounts of FA in the initial sample. The amounts should be multiplied by 2.5 (= 1.0/0.4) for samples prepared using Procedure C (in which 0.4 ml from the total 1.0 ml was analyzed).
Notes
With the exception of pipette tips, do not use plasticware (tubes and containers) for organic solvents, as the quantification of several FAMEs will fail due to components leaching from the plasticware. For the same reason, do not transfer reagents from the Fatty Acid Methylation Kit into plastic containers before use. To volumetrically measure organic solvents accurately, pipettes should be pre-wetted with the solvent until no further drips are observed. This takes more than 10 aspiration/ejection cycles.
For safety, organic solvents should be used in a fume hood or a draught chamber. Wear lab coats and use protective glasses and gloves.
Recipes
- Chloroform:2-propanol (2:1, v:v)
Note: Prepare when required.
Separately measure 40 ml of chloroform and 20 ml of 2-propanol with a glass graduated cylinder and mix in a clean glass bottle - 2% acetic acid in diethyl ether
Note: Prepare when required.- Measure 60 ml of diethyl ether using a graduated cylinder
- Transfer the diethyl ether to a clean glass bottle
- Add 1.2 ml of acetic acid with a Pipetman (P1000 or P5000)
- 2.8% ammonia in methanol
Note: Prepare when required.- Measure 54 ml of methanol using a graduated cylinder
- Transfer the methanol to a clean glass bottle
- Add 6 ml of 28% ammonia solution with a Pipetman (P1000 or P5000)
- PBS
Dilute 5 ml of PBS (10x conc.) with ultra-pure water to a total volume of 50 ml - C23:0 FA (5 mg/ml stock and 1 mg/ml working solutions)
- Prepare a chloroform:methanol (1:1, v:v) mixture in a similar manner to the chloroform:2-propanol (2:1) mixture, described above
- Weigh 20 mg of C23:0 FA powder using a balance and dissolve in 4 ml of chloroform:methanol (1:1, v:v) in a glass test tube (16 x 125 mm) using a pre-wetted pipette
- To prepare the 1 mg/ml solution, dilute 1 ml of the 5-mg/ml stock solution with 4 ml of methanol in a glass tube (16 x 125 mm)
- Both stock and working solutions can be stored at -30 °C in 2.0-ml autosampler vials sealed with screw caps
- Standard FAME mixtures (serial dilution)
Note: Prepare when required and use a pre-wetted pipette for quantitative accuracy.- Label four empty 0.5-ml microtubes as ‘std1’, ‘std2’, ‘std3’, and ‘std4’.
- Place 30 µl of the Supelco 37-component FAME mix in the std1 tube. Dispense 30 µl of dichloromethane into the std2, std3, and std4 tubes. Use a pre-wetted pipette for accuracy.
- Transfer 10 µl of std1 into std2 with a pre-wetted pipette and vortex (1/4 diluted).
- Repeat the four-fold serial dilution: std2 → std3 (1/16 diluted) and std3 → std4 (1/64 diluted).
- Transfer std1 to std4 to glass GC vial inserts. Label the GC vials as ‘std1’ to ‘std4’. The concentrations of the FAME standards in each vial are listed in Table 2.
Note: Do not store the std1 to std4 tubes for extended times as they will become contaminated with plasticware components.
- DTA (C22:4n-6) FAME (~1 mg/ml stock solution)
Evaporate ~20 µl of a 50-mg/ml solution of C22:4n-6 FAME in ethanol to dryness under a gentle stream of nitrogen gas and reconstitute with 1 ml of dichloromethane. Store at -30 °C - DPA (C22:5n-6) FAME (~10 mg/ml and ~1 mg/ml stock solutions)
- Add 1 ml of dichloromethane to C22:5n-6 in a glass ampule containing ~10 mg of C22:5n-6 methyl ester and transfer it to a clean glass vial (~10 mg/ml stock).
- Dilute the ~10 mg/ml stock with dichloromethane to prepare a ~1 mg/ml solution.
- Store at -30 °C
- DPA (C22:5n-3) FAME (~1 mg/ml stock solution)
- Evaporate ~100 µl of a 10-mg/ml solution of C22:5n-3 FAME in heptane to dryness under a gentle stream of nitrogen gas and reconstitute with 1 ml of dichloromethane
- Store at -30 °C
- DTA and DPA FAME mix
- Mix equal volumes (100 µl each) of the C22:4n-6, C22:5n-6, and C22:5n-6 FAME solutions in a 0.5-ml microtube
- Store at -30 °C
Acknowledgments
This work was supported by the AMED Practical Research Project for Allergic Diseases and Immunology (Research on Allergic Diseases and Immunology) 19ek0410040h0003 (to YK), AMED-CREST 19gm0910011 (to HS), AMED-P-CREATE 19cm0106116 (to HS), and the AMED Program for Basic and Clinical Research on Hepatitis JP19fk0210041 (to HS). TS was supported by the Takeda Science Foundation. This protocol was derived from our original work (Hashidate-Yoshida et al., 2015).
Competing interests
The Department of Lipidomics conducts joint research with the Shimadzu corporation. The Department of Lipid Signaling, National Center for Global Health and Medicine, conducts joint research with the Ono pharmaceutical company. The authors declare no other competing interests.
References
- Bligh, E. G. and Dyer, W. J. (1959). Canadian Journal of Biochemistry and Physiology. Can J Biochem Physiol 37(8).
- Egberts, J. and Bulskool, R. (1988). Isolation of the acidic phospholipid phosphatidylglycerol from pulmonary surfactant by sorbent extraction chromatography. Clin Chem 34(1): 163-164.
- Hashidate-Yoshida, T., Harayama, T., Hishikawa, D., Morimoto, R., Hamano, F., Tokuoka, S. M., Eto, M., Tamura-Nakano, M., Yanobu-Takanashi, R., Mukumoto, Y., Kiyonari, H., Okamura, T., Kita, Y., Shindou, H. and Shimizu, T. (2015). Fatty acid remodeling by LPCAT3 enriches arachidonate in phospholipid membranes and regulates triglyceride transport. Elife 4: 06328.
- Hishikawa, D., Hashidate, T., Shimizu, T. and Shindou, H. (2014). Diversity and function of membrane glycerophospholipids generated by the remodeling pathway in mammalian cells. J Lipid Res 55(5): 799-807.
- Hui, D. Y. and Howles, P. N. (2005). Molecular mechanisms of cholesterol absorption and transport in the intestine. Semin Cell Dev Biol 16(2): 183-192.
- Ichihara, K. and Fukubayashi, Y. (2010). Preparation of fatty acid methyl esters for gas-liquid chromatography. J Lipid Res 51(3): 635-640.
- Ichihara, K., Shibahara, A., Yamamoto, K. and Nakayama, T. (1996). An improved method for rapid analysis of the fatty acids of glycerolipids. Lipids 31(5): 535-539.
- Iizuka-Hishikawa, Y., Hishikawa, D., Sasaki, J., Takubo, K., Goto, M., Nagata, K., Nakanishi, H., Shindou, H., Okamura, T., Ito, C., Toshimori, K., Sasaki, T. and Shimizu, T. (2017). Lysophosphatidic acid acyltransferase 3 tunes the membrane status of germ cells by incorporating docosahexaenoic acid during spermatogenesis. J Biol Chem 292(29): 12065-12076.
- Liu, K. S. (1994). Preparation of fatty acid methyl esters for gas-chromatographic analysis of lipids in biological materials. J Am Oil Chem Soc 71(11):1179-1187.
- Nielsen, T. S., Jessen, N., Jørgensen, J. O., Møller, N. and Lund, S. (2014). Dissecting adipose tissue lipolysis: molecular regulation and implications for metabolic disease. J Mol Endocrinol 52(3): R199-222.
- Shindou, H., Koso, H., Sasaki, J., Nakanishi, H., Sagara, H., Nakagawa, K. M., Takahashi, Y., Hishikawa, D., Iizuka-Hishikawa, Y., Tokumasu, F., Noguchi, H., Watanabe, S., Sasaki, T. and Shimizu, T. (2017). Docosahexaenoic acid preserves visual function by maintaining correct disc morphology in retinal photoreceptor cells. J Biol Chem 292(29): 12054-12064.
- Van Meer, G., Voelker, D. R., and Feigenson, G. W. (2008). Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol 9(2):112–24.
- Vorbeck, M. L., Mattick, L. R., Lee, F. A., and Pederson, C. S. (1961). Preparation of Methyl Esters of Fatty Acids for Gas-Liquid Chromatography: Quantitative Comparison of Methylation Techniques. Analytical Chemistry 33(11):1512-1514.
Article Information
Copyright
Hamano et al. This article is distributed under the terms of the Creative Commons Attribution License (CC BY 4.0).
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Hamano, F., Tokuoka, S. M., Hashidate-Yoshida, T., Shindou, H., Shimizu, T. and Kita, Y. (2020). Quantification of Fatty Acids in Mammalian Tissues by Gas Chromatography–Hydrogen Flame Ionization Detection. Bio-protocol 10(9): e3613. DOI: 10.21769/BioProtoc.3613.
- Hashidate-Yoshida, T., Harayama, T., Hishikawa, D., Morimoto, R., Hamano, F., Tokuoka, S. M., Eto, M., Tamura-Nakano, M., Yanobu-Takanashi, R., Mukumoto, Y., Kiyonari, H., Okamura, T., Kita, Y., Shindou, H. and Shimizu, T. (2015). Fatty acid remodeling by LPCAT3 enriches arachidonate in phospholipid membranes and regulates triglyceride transport. Elife 4: 06328.
Category
Biochemistry > Other compound > Acid
Biochemistry > Lipid > Lipid measurement
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









