- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Application of Mechanical Forces on Drosophila Embryos by Manipulation of Microinjected Magnetic Particles
Published: Vol 10, Iss 9, May 5, 2020 DOI: 10.21769/BioProtoc.3608 Views: 4396
Reviewed by: Samantha E. R. DundonImre GáspárTrinadh Venkata Satish Tammana

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Labeling and Tracking Mitochondria with Photoactivation in Drosophila Embryos
Sayali Chowdhary and Richa Rikhy
Mar 5, 2022 4329 Views

Measurement of Contractile Ring Tension Using Two-photon Laser Ablation during Drosophila Cellularization
Swati Sharma and Richa Rikhy
Mar 20, 2022 2498 Views

Protocol for Imaging the Same Class IV Neurons at Different Stages of Development
Sonal Shree and Jonathon Howard
Aug 20, 2024 1318 Views
Abstract
Cells generate mechanical forces to shape tissues during morphogenesis. These forces can activate several biochemical pathways and trigger diverse cellular responses by mechano-sensation, such as differentiation, division, migration and apoptosis. Assessing the mechano-responses of cells in living organisms requires tools to apply controlled local forces within biological tissues. For this, we have set up a method to generate controlled forces on a magnetic particle embedded within a chosen tissue of Drosophila embryos. We designed a protocol to inject an individual particle in early embryos and to position it, using a permanent magnet, within the tissue of our choice. Controlled forces in the range of pico to nanonewtons can be applied on the particle with the use of an electromagnet that has been previously calibrated. The bead displacement and the epithelial deformation upon force application can be followed with live imaging and further analyzed using simple analysis tools. This method has been successfully used to identify changes in mechanics in the blastoderm before gastrulation. This protocol provides the details, (i) for injecting a magnetic particle in Drosophila embryos, (ii) for calibrating an electromagnet and (iii) to apply controlled forces in living tissues.
Keywords: Drosophila embryosBackground
Drosophila melanogaster embryogenesis is a classical model for morphogenesis (Campos-Ortega and Hartenstein, 1985). While many tools have been developed to assess the role of specific proteins in morphogenesis, assessing cellular forces or mechanics still remains challenging. For the last fifteen years, laser dissection has been the most commonly used approach to assess cellular forces (Colombelli and Solon, 2013; Shivakumar and Lenne, 2016). However, laser dissection is invasive, it wounds tissues, and does not facilitate the application of ectopic forces. To overcome these limitations, a few methods have been developed to probe the mechanics of tissues by inducing deformations in droplets of magnetic fluid or through the optical trapping of cellular junctions (Bambardekar et al., 2015; Serwane et al., 2017). These methods have a limited range of force and require specific, complex instrumentation. Here, we present the protocol for an alternative, versatile and low-cost method for applying controlled forces within an epithelium of a living Drosophila embryo (D’Angelo et al., 2019). This method relies on the injection of a magnetic particle within a living embryo and on the application of a magnetic field with an electromagnet. Since the magnetic particle is coated with GBP nanobody, it is possible to target a specific intracellular attachment. Here, we injected the bead in a fly line expressing GFP at the plasma membrane (Resille GFP) to position the bead at the plasma membrane. Our methodology does not impair morphogenesis and repeated force application can be performed without cellular damage.
It is well established that tissue mechanics is an essential component to consider for the control of cellular behavior (differentiation, cell division, migration…) leading to morphogenesis during animal development and to the progression of diseases such as tumor formation and cancer progression (Lecuit et al., 2011; Heisenberg and Bellaiche, 2013; Engler et al., 2006; Frey et al., 2008; Godard and Heisenberg, 2019; Northcott et al., 2018). In these contexts, our method can be used to probe tissue mechanics and its changes or to apply local forces to investigate mechanosensation and mechanotransduction.
Materials and Reagents
- 1.5 ml Eppendorf tube
- Magic tape (Scotch)
- Glass capillaries 1 mm in diameter, 90 mm length (Narishige model G1)
- Cover slip 24 x 60 mm, thickness 1
- FrameSlides (Leica microsystem, catalog number: 11505151)
- Silicon grease (Kluber lubrication, catalog number: 0040260221)
- Neodymium magnet (RS, catalog number: 695-0169)
- Parafilm (VWR, catalog number: 291-1214)
- Mu-metal rod 100 x 4 mm (Sekels)
- Aluminium cylinder (45 mm x 200 mm)
- Copper wire 0.5 mm
- Drosophila melanogaster embryos expressing Resille GFP
Note: This can be easily extended to other Drosophila line or organism. - Dynabeads M-450 tosylactivated (Thermo Fisher, catalog number: 14013)
- GFP binding protein (GBP) custom made (D'Angelo et al., 2019)
Note: The GBP can be substituted by commercial anti GFP or other types of antibodies. - Boric Acid (Sigma-Aldrich, catalog number: B6768)
- PBS
- PDMS (Poly(dimethylsiloxane), hydroxy terminated 750 cSt (Sigma-Aldrich, catalog number: 481963)
- Heptane (Sigma-Aldrich, catalog number: H2198)
- Sodium Hypochlorite (Panreac, catalog number: 2119211211)
- Voltalef oil 10s (VWR, catalog number: 9002-83-9)
- BSA (Bovine Serum Albumin) (Sigma-Aldrich, catalog number: A2153)
Equipment
- Microinjection microscope (Leica, model: DMIL ) equipped with 20x objective and a micromanipulator (Narishige, model: MN-153 )
- Micro-puller (Sutter instruments, model: P30 ) equipped with a 3 mm wide Trough filament (Sutter Instrument, catalog number: FT00330B )
- Microinjector (WPI, model: PV 820 )
- Micro grinder (Narishige, model: EG-44 )
- Andor Revolution XD spinning disk confocal microscope equipped with 40x and 100x objectives and lasers emitting at 488 nm and 561 nm
- Three-axis micromanipulator (Narishige, model: UMM-3FC )
- Bioruptor (Diagenode, model: UCD-200TM-EX )
- Thermomixer (Eppendorf, model: 5350 )
- Hot plate (Stuart, model: US152 )
- Power supply 30 V, 5A (Blausonic, model: FA-350 )
Software
- Fiji (Schindelin et al., 2012)
- Excel (or alternative plotting and fitting software)
Procedure
- Construction and calibration of the electro magnet
To apply controlled forces, we designed an electromagnet similar to the one described in Kollmansberger and Fabry (Kollmannsberger and Fabry, 2007). The electromagnet is composed of three components: A Mu-metal rod (soft ferromagnetic alloy with very high magnetic permeability), a radiator in aluminium to evacuate the heat and a copper wire to connect to electric current (Figures 1A and 1B). The Mu-metal rod is sharpened to achieve a conical tip and then thermally annealed. (both operations can be done at the Sekels factory). A custom manufactured 45 mm long aluminium cylinder–with 4.5 mm inner diameter to host the Mu-metal rod was used to dissipate the heat generated by the magnet. The outer diameter of the cylinder was 200 mm. Close to one end, four, 1 mm wide and 40 mm deep troughs spaced by 2 mm were milled into the cylinder to increase the radiator surface. Another 220 mm wide trough of 100 mm diameter was milled 5 mm from the other end of the cylinder to host 100 coils of the copper wire. On the two extremities, three plastic screw were positioned at 120° interval and were used to hold the Mu-metal rod (Figures 1C and 1D).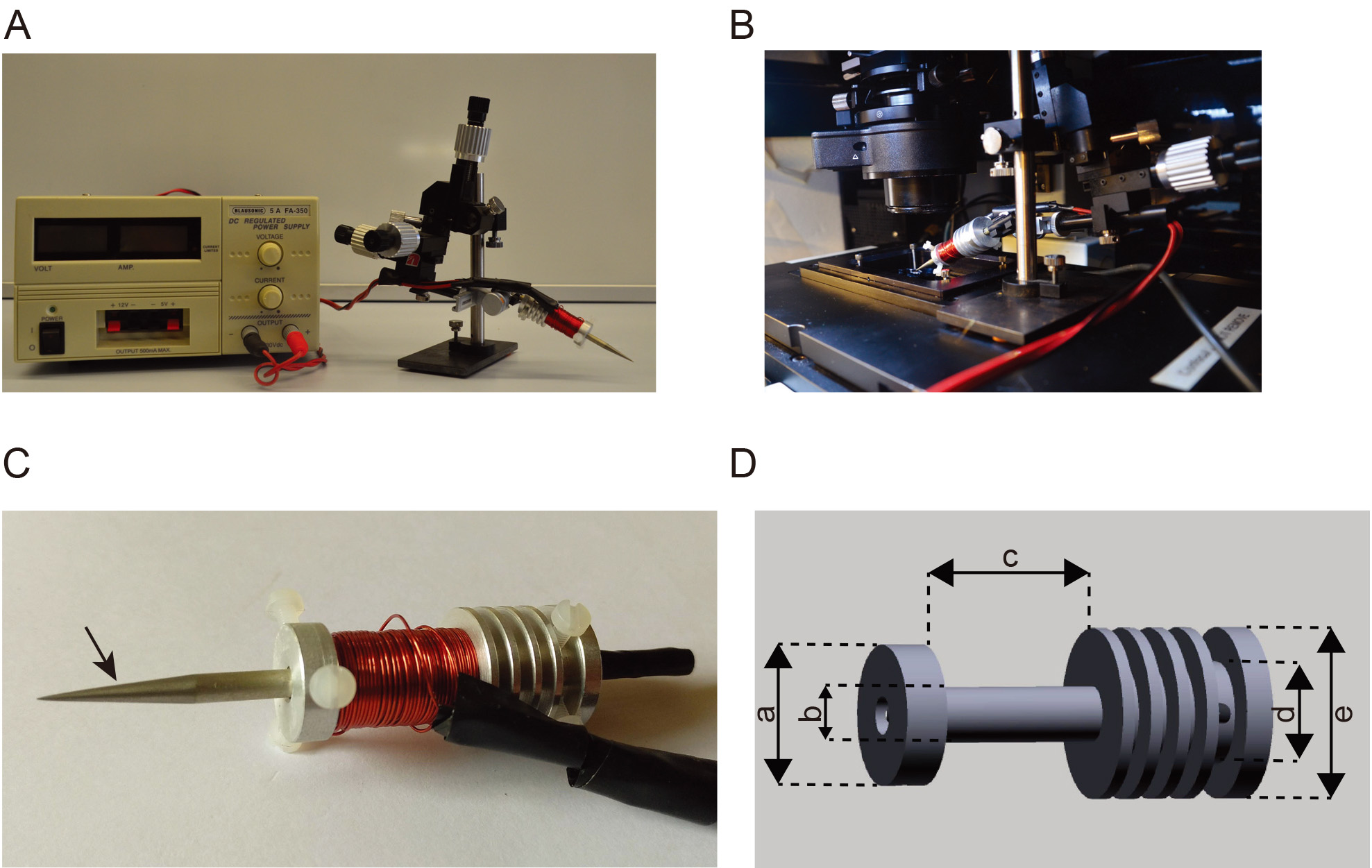
Figure 1. Electromagnet set up for force application. A. Photograph of the power supply (left) and electromagnet mounted on the micromanipulator. B. Photograph of the electromagnet mounted on the stage of the spinning disk microscope. C. Close up of the electromagnet, the left arrow indicates the Mu-metal rod conically shaped, the center arrow indicates the copper coil around the radiator and the right arrow indicates the aluminium radiator. D. Schematics showing the design of the aluminium radiator, a = 160 mm, b = 120 mm, c = 220 mm, d = 160 mm, e = 200 mm.
Upon application of a direct current circulating within the cupper coils, a magnetic field emanates from the tip and decays spatially. Magnetic beads lying within the magnetic field are attracted towards the tip with a force proportional to the gradient of the field (Kollmannsberger and Fabry, 2007).
To calibrate the force exerted on the bead as a function of the distance from the magnet, we developed an assay using beads embedded in PDMS. Since its viscosity is known, by measuring the velocity of the beads within the PDMS when the magnetic field is applied, we can calculate the force field as a function of the distance from the tip: Stoke’s law directly relates the force to the velocity (Figure 2 and Video 1).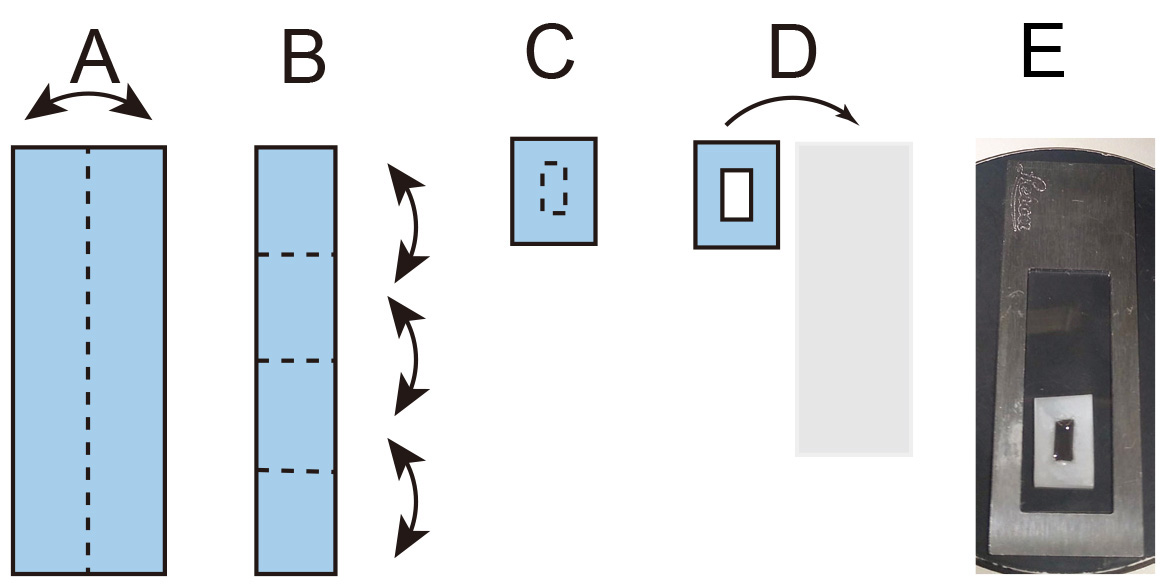
Figure 2. Construction of the calibration chamber. A and B. Sequential folding of a rectangular piece of Parafilm, dotted line represents the folding site. C. The dotted line represents the cut position to generate a window. D. The Parafilm is transferred onto a coverslip. E. Photograph of the calibration chamber mounted on the metal slide.Video 1. Calibration of the force applied to a 4.5 μm bead. (Left) Time lapse video of 4.5 μm magnetic beads embedded in PDMS moving toward the magnet during application of the magnetic field and respective tracking (Right).- Place 5 μl of Dynabeads in 1.5 ml Eppendorf tube and put it at 37 °C with the lid open in the Thermomixer for a few hours in order to completely desiccate the bead.
- Add 1 ml of PDMS and stir it with a pipette tip in order to detach the pellet of beads from the bottom.
- Using the Bioruptor, sonicate at maximum power for 10 min. If the beads are not evenly resuspended repeat the sonication procedure.
- Cut a rectangular piece of Parafilm 10 cm x 3 cm and fold it as in Figure 2.
- Using a cutter, open a window of 5 x 10 mm in the center of the folded Parafilm.
- Transfer the Parafilm on top of a coverslip and heat it on top of a hot plate until the Parafilm attaches properly to the coverslip without melting.
- Transfer 50 μl of beads/PDMS (from Step A3) into the Parafilm window.
- Mount the coverslip onto a frame slide Leica using a drop of silicon grease.
- On the spinning disk microscope use a 40x lens and focus on the beads.
- Bring the electromagnet to one edge of the field of view (Video 1).
- Start the time-lapse using transmitted light and an exposure time of 20 ms.
- Start the force application.
- Injection procedure of individual 4.5 μm bead into Drosophila embryo
A single magnetic bead is injected into the yolk of an embryo at stage 2 of Drosophila development (Figures 3-4). After injection, the coverslip supporting the embryos is left to develop over a neodymium magnet that will hold the bead in the embryo cortex. Thus, at cellularization time, the bead will be naturally encapsulated into a newly formed cell. By choosing the orientation of the embryo relative to the neodymium magnet, it is possible to embed the particle in a specific group of cells, which will later develop into specific tissues or organs.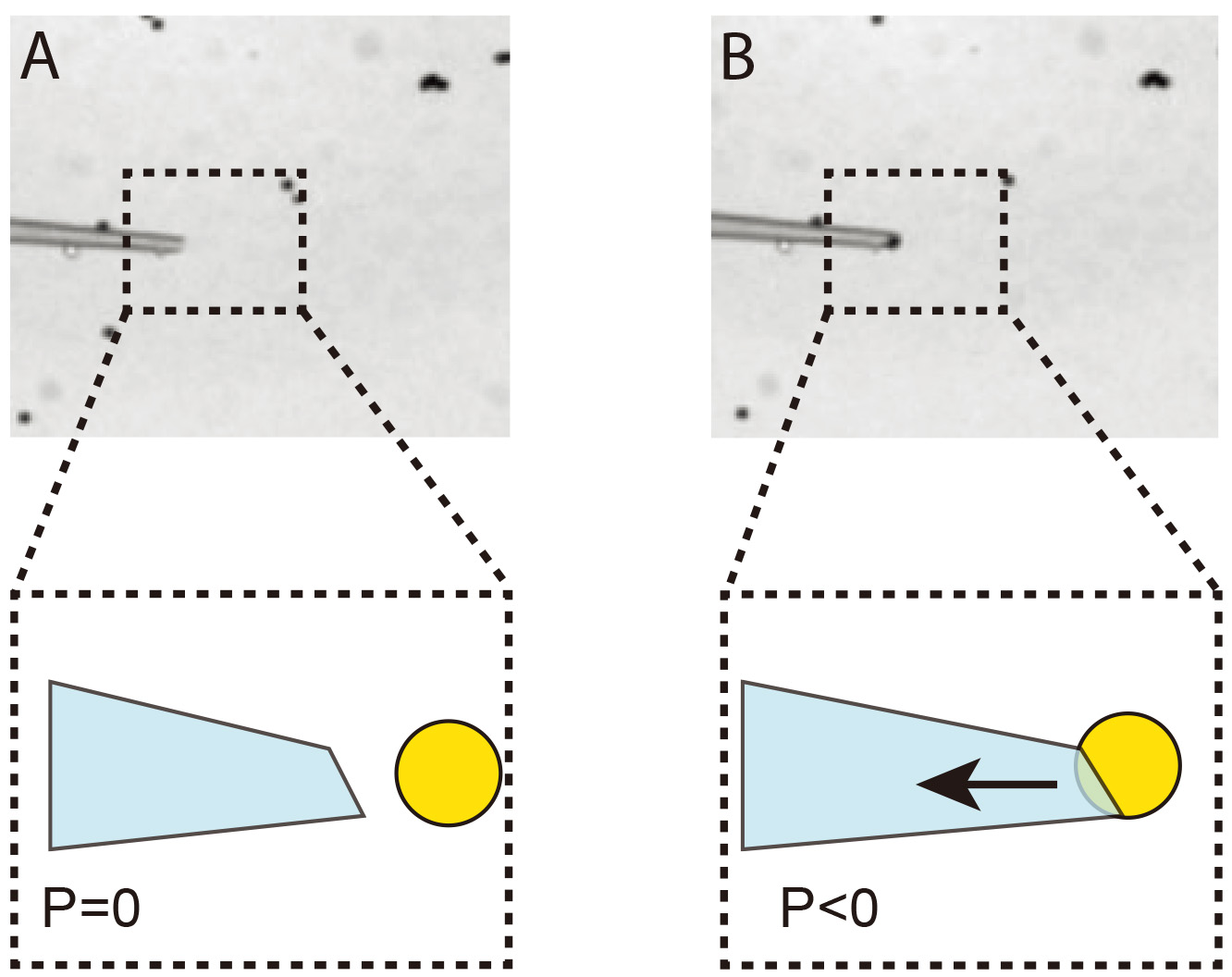
Figure 3. Immobilization of a single 4.5 μm bead at the tip of the injection needle. A. The injection needle is placed into a drop of 4.5 μm beads resuspended in 0.1% PBS/BSA. B. Negative pressure (P < 0) is applied to the needle in order to immobilize a single bead.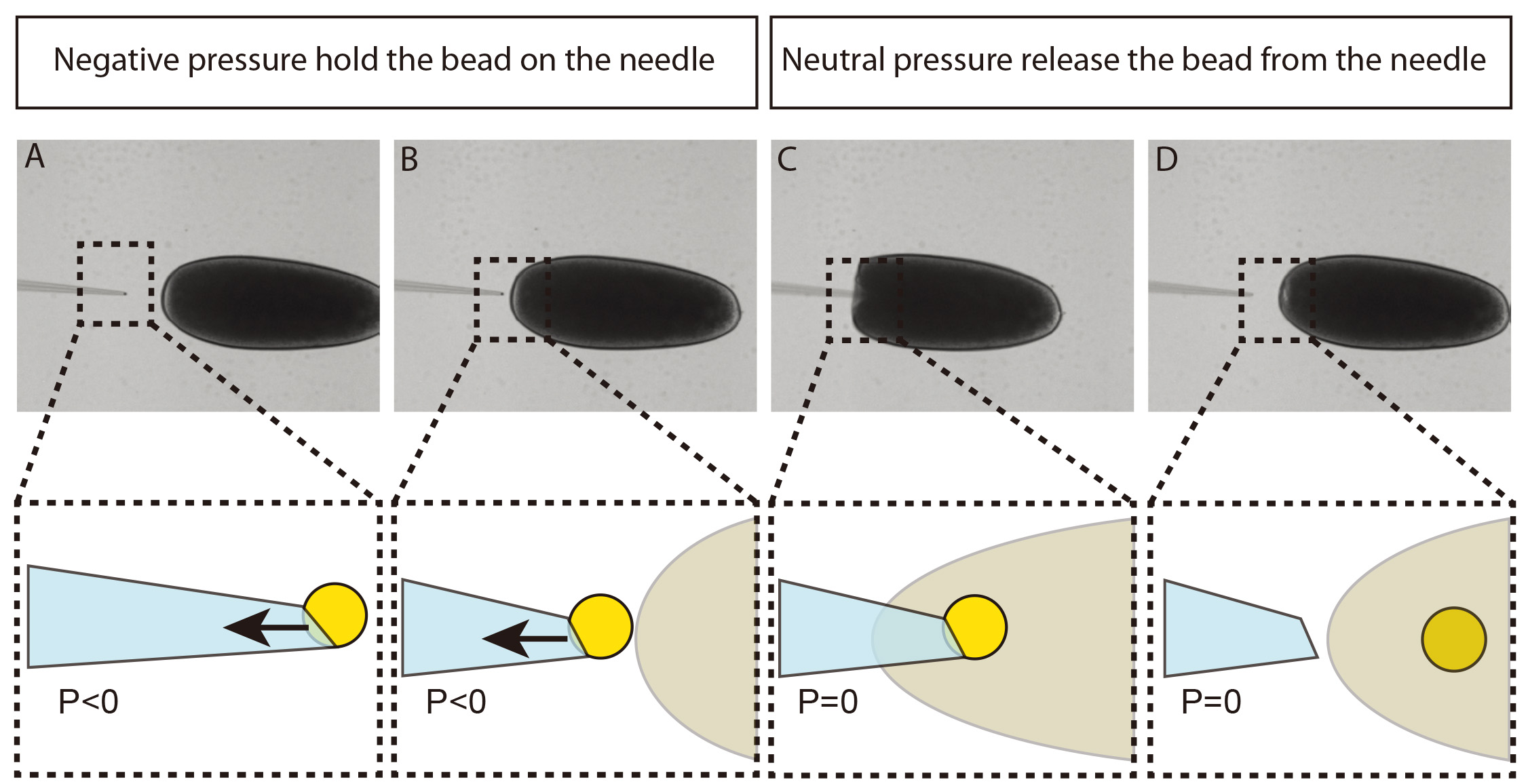
Figure 4. Injection of a single 4.5 μm bead into Drosophila embryo. A and B. Negative pressure (P < 0) is applied to the needle in order to immobilize a single bead while approaching the embryo. C and D. Negative pressure is released and the bead remains inside the embryo. The inset shows a cartoon of the corresponding injection step where the needle is depicted in cyan, the bead in yellow and the embryo in light gray. The arrow represents the resulting force generated by the negative pressure inside the needle.- Prepare 0.1 M boric acid solution at pH 9.5.
- Wash 3 times 50 μl of 4.5 μm Dynabeads with 1 ml of boric acid solution.
- Resuspend 10 μg of GBP in 400 μl of boric acid solution and incubate O/N at 37 °C.
- Wash 2 times the Dynabeads with 1 ml of boric acid solution.
- Resuspend in 1 ml boric acid solution 0.1% BSA (w/v).
- Prepare heptane glue by incubating O/N 10 ml of heptane with 120 cm of magic tape.
- Place a drop of 100 μl of heptane glue in the middle of a coverslip and allow the drop to flow down by gravity, creating a stripe of sticky surface in the middle of the coverslip, parallel to his long edge.
- Thirty to forty freshly laid embryos are chemically dechorionated using sodium hypochlorite for 90 s at RT, then rinsed under tap water for 90 s and then mounted on a coverslip coated with heptane glue.
Note: All embryos should be mounted in a single row, regularly spaced and parallel to each other, to facilitate the injection. - The embryos are dehydrated for 10-15 min in a 25 °C incubator, to reduce turgidity and avoid bursting upon injection, and then covered with a small drop of Voltalef oil. Please note that the dehydration time needs to be adjusted depending on the humidity in the incubator.
- A drop of 40 μl beads resuspended in PBS/0.1% BSA is deposited next to the embryos, aside of the Voltalef oil drop.
- Turn on the micro-puller and set the values of heat 1 at 860 and pull at 750. Then insert a glass capillary in the respective holder and generate an injection needle. Note that different pulling parameters can be used. The injection needle just need to be elongated enough to have a diameter allowing to hold the magnetic particle and to enter the embryo without damaging it (see Figures 3 and 4).
- The injection needle is precisely opened using a microgrinder to achieve an internal diameter of 3.5 μm (see Figure 3).
- The needle is connected to a Microinjector (WPI PV 820 ).
- The needle is positioned in the drop containing beads (in PBS/0.1% BSA) and, by applying negative pressure, a single bead is held at the tip of the needle. Figure 3 illustrates the process.
- Move the needle holding the bead close to the posterior pole of the embryo, set the pressure of the microinjector to neutral just before the injection. Then, the bead can be transferred into the center of the embryo (see Figure 4).
- Once all the embryos are injected, place the coverslip with the injected embryos on top of a permanent magnet for 2 h at 25 °C.
- In order to maximize the force applied by the neodymium magnet to the embryos, position the coverslip with the injected embryos directly on the neodymium magnet without the microscopy slide.
- To minimize the damage inferred to the embryos, the bead needs to be injected at early developmental stage (stage 2). In order to target specific tissues, the embryo needs to be mounted with the tissue of interest facing the coverslip. i.e., if we want to apply forces during the process of dorsal closure, the embryos have to be mounted with dorsal side facing the coverslip and the embryos must be left to develop for 20 h after injection at 18 °C.
- Imaging and force application
- The coverslip is mounted onto a frame slide Leica using a drop of silicon grease.
- Select a 40x lens at the spinning disk, focus on an embryo with the bead in right orientation and then position the tip of the electro-magnet at 150-200 μm from the chosen embryo. Care must be taken in choosing the bead-magnet distance in order to position the bead in a region of the force-distance curve with small steepness to allow for an accurate estimation of the force (see Figure 5B and Figure 6).
- Set up a two channel, x-y-z time lapse imaging protocol. One channel will use the 488 nm laser to acquire GFP signal and the other channel will use 561 nm laser will acquire the auto-fluorescence of the bead.
- Set up 5 z steps spaced by 1 μm and choose a time resolution of 5 s.
- Start the movie, after 1 min turn on the power supply and select 0.3 A to generate a magnetic field that will exert a force of 115pn on the bead. Keep the power supply on for 1 min, then, after switching off the power supply, allow the imaging to proceed for 1 more minute before stopping the movie (Figures 5A, 5C and Video 2).
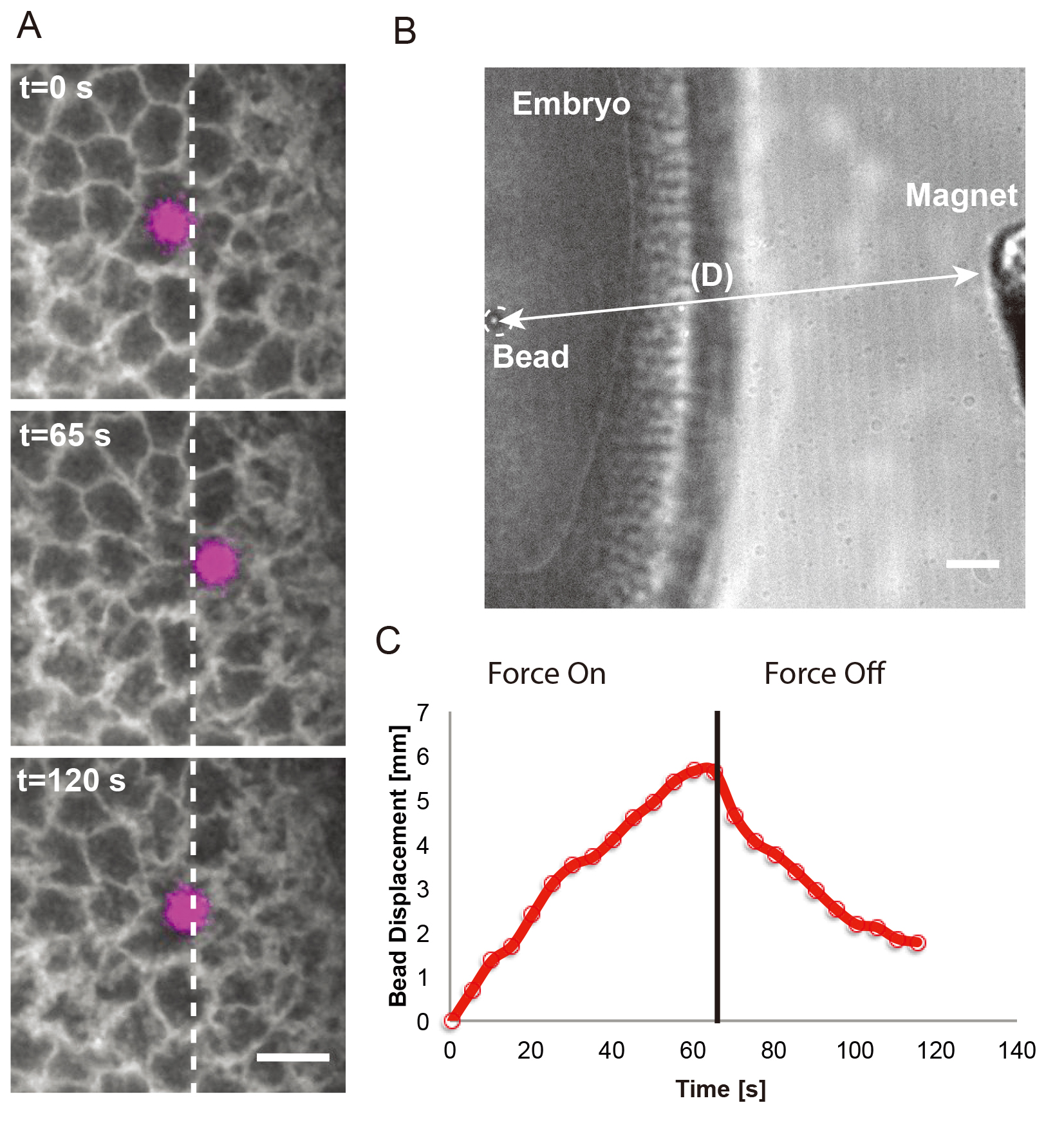
Figure 5. Force application on a single 4.5 μm bead to the Drosophila blastoderm. A. Time-lapse images showing the bead (magenta) in response to a force step (∼115 pN; onset at 0 s). The bead was embedded into an individual cell of a Resille-GFP embryo. White arrows indicate force application. White dashed lines mark the left side of the bead at time 0s. Scale bar = 10 μm. B. Snapshot in transmitted light of an embryo containing a 4.5 μm bead and the electromagnet placed at 190 μm. Scale bar = 20 μm. C. Bead displacement corresponding to the force application shown in A.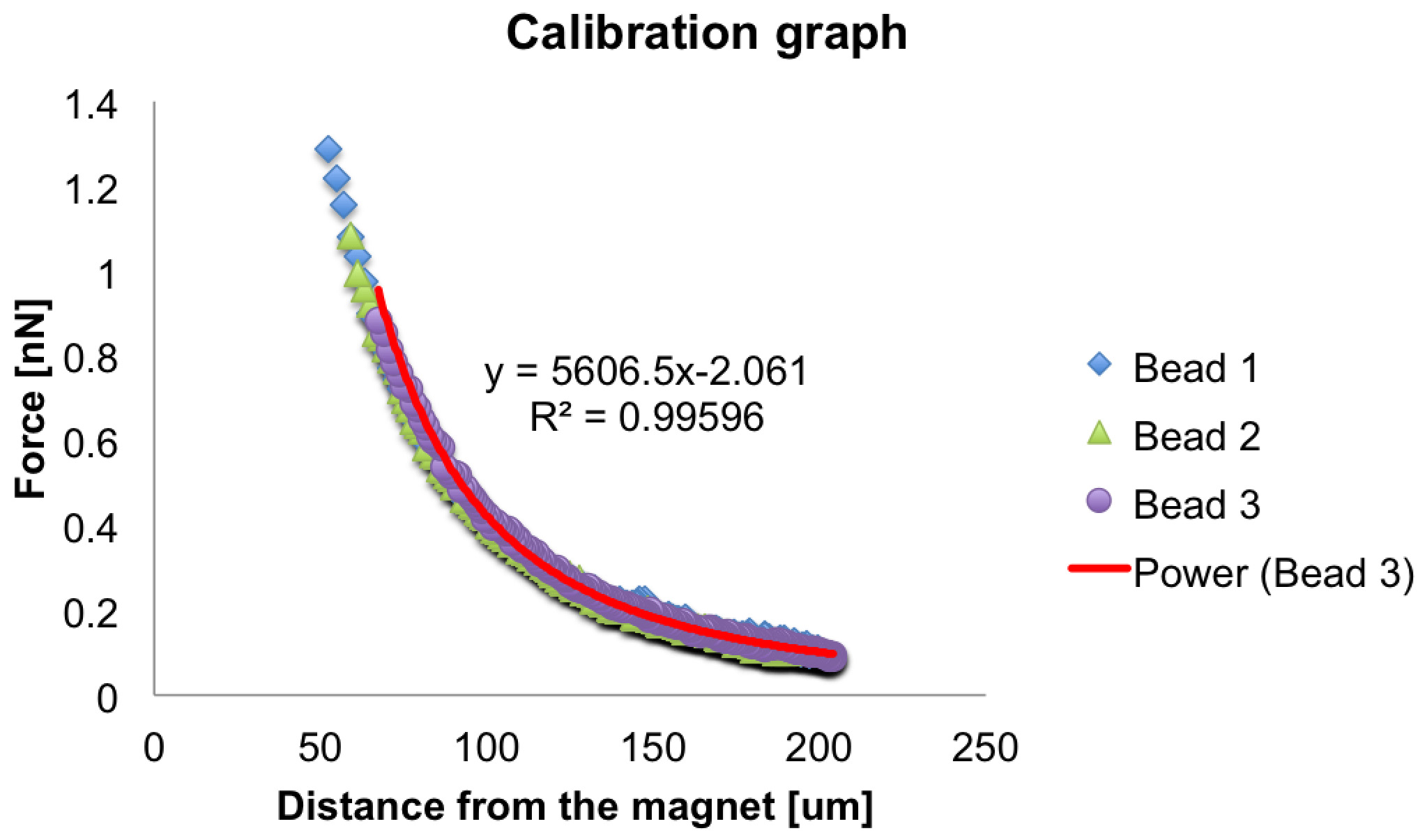
Figure 6. Force-distance calibration plot. Example of force-distance plot for the calibration of the electromagnet. The force exerted on three single beads is plotted as function of the distance from the tip of the magnet. A power law fit on the force curve of bead number three is represented in red.Video 2. Force delivery to a single cell during Drosophila cellularization. Time-lapse video showing a force application on a 4.5 μm bead embedded into a Resille-GFP expressing embryo. The purple arrow indicates when a force of ~115 pN is applied on the bead. Scale bar = 10 μm.
Data analysis
- Force Calibration
- Open the time-lapse performed during the calibration with Fiji. Apply a median filter (2 pixels) to reduce the noise.
Note: Other types of filter (like Gaussian blur filter for instance) can be used, the only criterion is to reduce sufficiently the background noise to correctly threshold the beads. - Threshold to achieve a binary movie, and convert it to 8 bit.
- Remove the scale of the movie
- Use the MTrack2 plugins to extract the X and Y coordinates of the beads during the force application (Video 1). Define the following parameters in the window associated to the plugin:
Minimum object size to 8 μm2 (corresponding to 20 pixels in our set up)
Maximum object size to 120 μm2 (corresponding 300 pixels in our set up)
Maximum velocity to 100
Minimum track length (frames) to 20
Tick Save Results File box
Tick Display Path Lengths box
Tick Show Labels box
Tick Show Positions box
Tick Show Paths box - The MTrack2 plugin generates a trackresult file with X1 and Y1 coordinates of the tracked objects at each time point.
- Import the trackresult file in Excel or other data plotting software. If the detection is noisy, one can perform a time average of few consecutive data points into new X and Y columns to smooth the trajectories.
- From the X and Y coordinates, we calculate the distance D of the bead from the magnet at each time point t.
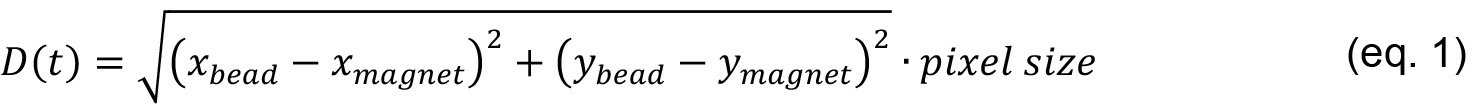
where,
pixel size is the size of a pixel in µm,
xbead is the x value (in pixels) of the bead at each time point,
ybead is the y value (in pixels) of the bead at each time point,
xmagnet is the x value (in pixels) of the magnet,
ymagnet is the y value (in pixels) of the magnet. - Calculate the velocity of the bead v(t)
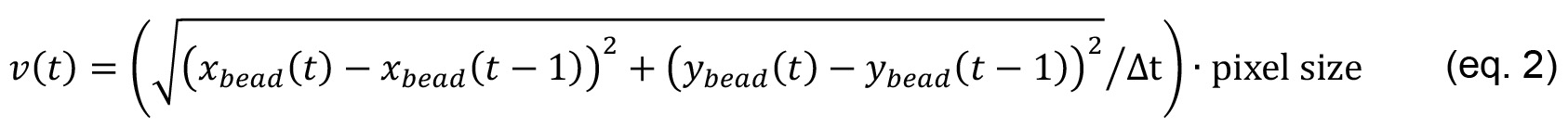
where,
Δt is the time interval of the movie in seconds. - The force F (t) applied on the bead can then be calculated at any time point using the following Stoke’s law.

where,
µ is the viscosity of the media in which the bead is embedded,
R is the radius of the bead,
v(t) is the velocity of the bead. - From the obtained values of D(t) and F(t), generate a scatter plot of the force as a function of the respective distance in the plotting software of your choice and then fit the resulting scatter plot with a power law with the following shape:

where A and α are fitting parameters (see Figure 6).
Note: That each individual bead track can be fitted independently and the extracted fitting parameters will then be averaged or a single fit of all the tracks could be generated.
- Open the time-lapse performed during the calibration with Fiji. Apply a median filter (2 pixels) to reduce the noise.
- Bead displacement analysis after force application in the blastoderm.
- Open the force application to blastoderm movie with Fiji and split the green (488 nm) and red (561 nm) channels (see Video 2).
- Sum project all the single z planes into single movie and then apply median filter (with a kernel of 2 pixels).
- Apply the steps 2 and 3 as in the Force Calibration section to the red channel.
- Folllow the step 4 as in the Force Calibration section but using the following parameters:
Minimum object size to 32 µm2 (corresponding 200 pixels in our set up)
Maximum object sixe to 400 µm2 (corresponding 2,500 pixels in our set up)
Maximum velocity to 100
Minimum track length (frames) to 20
Tick Save Results File box
Tick Display Path Lengths box
Tick Show Labels box
Tick Show Positions box
Tick Show Paths box - By applying equation 1 to the X and Y coordinates generated by the MTrack2 plugin, and replacing the coordinates of the magnet with the initial coordinates of the bead in the generated file, we can calculate the bead displacement during the force application at each time point t (Figure 6).
- To estimate the force applied to the bead we measure the distance D from the magnet to the initial position of the bead, again using the equation 1. Then, we calculate the force using the equation 4 with the parameters previously estimated during the calibration.
Acknowledgments
We thank Peran Hayes for discussions and critical reading of the manuscript and Roger Uceda from the Fundació CIM for manufacturing the aluminium radiator. We are grateful to the ALMU team for providing help with the microscopy. The research leading to these results has received funding from the Spanish Ministry of Economy and Competitiveness, Plan Nacional, BFU2010-16546 and BFU2015-68754, “Centro de Excelencia Severo Ochoa” and to the EMBL partnership. We acknowledge the support of the CERCA Programme/Generalitat de Catalunya. This work was supported in part by the Fundaciòn Biofisika Bizkaia and the Basque Excellence Research Centre (BERC) program of the Basque Governement.
Competing interests
There are no conflicts of interest or competing interest.
References
- Bambardekar, K., Clement, R., Blanc, O., Chardes, C. and Lenne, P. F. (2015). Direct laser manipulation reveals the mechanics of cell contacts in vivo. Proc Natl Acad Sci U S A 112(5): 1416-1421.
- Campos-Ortega, J. A., and Hartenstein, V. (1985). The embryonic development of Drosophila melanogaster. Berlin, Springer-Verlag.
- Colombelli, J. and Solon, J. (2013). Force communication in multicellular tissues addressed by laser nanosurgery. Cell Tissue Res 352(1): 133-147.
- D'Angelo, A., Dierkes, K., Carolis, C., Salbreux, G. and Solon, J. (2019). In vivo force application reveals a fast tissue softening and external friction increase during early embryogenesis. Curr Biol 29(9): 1564-1571 e1566.
- Engler, A. J., Sen, S., Sweeney, H. L. and Discher, D. E. (2006). Matrix elasticity directs stem cell lineage specification. Cell 126(4): 677-689.
- Frey, B., Janko, C., Ebel, N., Meister, S., Schlucker, E., Meyer-Pittroff, R., Fietkau, R., Herrmann, M. and Gaipl, U. S. (2008). Cells under pressure - treatment of eukaryotic cells with high hydrostatic pressure, from physiologic aspects to pressure induced cell death. Curr Med Chem 15(23): 2329-2336.
- Godard, B. G. and Heisenberg, C. P. (2019). Cell division and tissue mechanics. Curr Opin Cell Biol 60: 114-120.
- Heisenberg, C. P. and Bellaiche, Y. (2013). Forces in tissue morphogenesis and patterning. Cell 153(5): 948-962.
- Kollmannsberger, P. and Fabry, B. (2007). High-force magnetic tweezers with force feedback for biological applications. Rev Sci Instrum 78(11): 114301.
- Lecuit, T., Lenne, P. F. and Munro, E. (2011). Force generation, transmission, and integration during cell and tissue morphogenesis. Annu Rev Cell Dev Biol 27: 157-184.
- Northcott, J. M., Dean, I. S., Mouw, J. K. and Weaver, V. M. (2018). Feeling stress: The mechanics of cancer progression and aggression. Front Cell Dev Biol 6:17.
- Serwane, F., Mongera, A., Rowghanian, P., Kealhofer, D. A., Lucio, A. A., Hockenbery, Z. M. and Campas, O. (2017). In vivo quantification of spatially varying mechanical properties in developing tissues. Nat Methods 14(2): 181-186.
- Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., Preibisch, S., Rueden, C., Saalfeld, S., Schmid, B., Tinevez, J.Y., White, D. J., Hartenstein, V., Eliceiri, K., Tomancak, P. and Cardona, A. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods 9(7):676-82.
- Shivakumar, P. C. and Lenne, P. F. (2016). Laser ablation to probe the epithelial mechanics in Drosophila. Methods Mol Biol 1478: 241-251.
Article Information
Copyright
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
D’Angelo, A. and Solon, J. (2020). Application of Mechanical Forces on Drosophila Embryos by Manipulation of Microinjected Magnetic Particles. Bio-protocol 10(9): e3608. DOI: 10.21769/BioProtoc.3608.
Category
Developmental Biology > Morphogenesis > Cell structure
Cell Biology > Tissue analysis > Tissue culture
Cell Biology > Tissue analysis > Stiffness measurement
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









