- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
A Modified Semisolid Clonal Culture for Identification of B-1 and B-2 Progenitor Colony Forming Ability of Mouse Embryonic Hemogenic Endothelial Cells
Published: Vol 10, Iss 9, May 5, 2020 DOI: 10.21769/BioProtoc.3601 Views: 4188
Reviewed by: Giusy TornilloWoojong LeeAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Isolation and Co-culture of Paneth Cells and Intestinal Stem Cells
Ryosuke Isotani [...] Toshimasa Yamauchi
Sep 20, 2025 3460 Views

Efficient Fluorescent Labeling of Human Trophoblast Stem Cells via a CRISPR/Cas9-Mediated Knock-In Approach in a Safe Harbor Locus
Hengshan Zhang [...] Danny J. Schust
Jan 5, 2026 294 Views

Simple and Rapid Model to Generate Differentiated Endometrial Floating Organoids
Adriana Bajetto [...] Tullio Florio
Feb 5, 2026 93 Views
Abstract
The search for the origin of the first hematopoietic stem cells (HSCs) in the mouse embryo has been a hot topic in the field of developmental hematopoiesis. Detecting lymphoid potential is one of the supportive evidence to show the definitive hematopoietic activity of HSCs. However, the first B-lymphoid potential in the mouse embryos are reported to be biased to innate-like B-1 cell lineage that can develop from hemogenic endothelial cells (HECs) independently of HSCs. On the other hand, conventional adaptive immune B cells (B-2) cells are considered to be exclusively derived from HSCs. Therefore, segregating B-1 and B-2 progenitor potential is important to understand the developmental process of HSCs that are also produced from HECs through intermediate precursors referred to as pre-HSCs. Both HECs and pre-HSCs show endothelial surface phenotype and require stromal support to detect their hematopoietic activity. The method utilizing stromal cell culture followed by modified semisolid clonal culture enables us to detect the number of colony forming units for B-1/B-2 progenitors originally derived from HECs/pre-HSCs, which will reflect the potential of B-1 biased or multi-lineage repopulating HSCs.
Background
Semisolid clonal culture (methylcellulose colony forming assay) is a traditional method to detect the number of hematopoietic progenitor cells. One colony is considered to be derived from a single progenitor cell (clonal origin). Added cytokines play important roles in the formation of yield colonies. For example, Epo enhances the colony forming unit of erythrocytes (CFU-E) or the burst forming unit of erythrocytes (BFU-E), while G-CSF/GM-CSF will enhance CFU-G (Granurocytes), M (Macrophages) or GM (Granulo-macrophages). SCF stimulate hematopoietic progenitor activity. The formation of B-lymphocyte progenitor colonies from mouse BM requires SCF, IL-7 and Flt3-ligand. Importantly, this assay can detect only the activity of CD45+ (or CD41+ in case of embryo-derived cells) hematopoietic progenitor cells, thus, cannot detect the hematopoietic activity of CD45-CD144+ hemogenic endothelial cells (HECs) that produce various hematopoietic cells in the mouse embryo. The detection of hematopoietic activity of HECs heavily depends on the stromal support or organ culture method. Therefore, it is challenging to determine the number of lymphoid progenitors produced from HECs. Recently, Montecino-Rodriguez et al. (2016) developed a modified semisolid clonal culture using S-17 stromal cells to detect B-1 and B-2 potential of hematopoietic progenitors in embryonic (E) day 10 yolk sac cells and fetal liver cells.
B-1 progenitor cells are mainly detected in the fetal liver and neonatal BM. B-1 cells belong to innate-like B-lymphocytes that produce natural IgM antibodies, while conventional adaptive immune B cells are referred to as B-2 cells. Montecino-Rodriguez et al., identified B-1 cell-specific progenitors in the mouse fetal liver and neonatal BM as lin-IgM-CD19+B220- (CD19 single positive) cells whereas B-2 specific progenitors (Pro-B cell) are lin-IgM-CD19-B220+ (B220 single positive) cells. These two populations quickly become CD19+B220+ double positive in vitro culture, thus, it is difficult to determine if B cells produced from embryonic tissues in the co-culture with stromal cells belong to B-1 or B-2 cells. This modified semisolid clonal culture enables us to detect B-1 and/or B-2 progenitor colonies derived from HECs/pre-HSC by utilizing stromal cells that support B lymphopoiesis (Kobayashi et al., 2019). We used OP9 stromal cells to support B-lymphopoiesis in the semisolid clonal culture, with which we have been successful to induce B lymphocytes from HECs (Yoshimoto et al., 2011; Kobayashi et al., 2019). Since B-2 progenitor colony forming ability seems to be one of the HSC activity, this method may be utilized to evaluate the HSC activity derived from HECs/pre-HSCs in the mouse embryo.
Materials and Reagents
- T-25 culture Flask (Corning, catalog number: 353109 )
- 6-well culture plates (Corning, Costar, catalog number: 3516 )
- 96-well Flat culture plates (Corning, Costar, catalog number: 3595 )
- Falcon Round bottom polypropylene14 ml tube (Corning, catalog number: 352059 )
- 1.5 ml Eppendorf tube
- 35 mm Petri dish (Falcon, catalog number: 351008 )
- 15 mm Petri dish
- 3 ml syringe (BD, catalog number: 309657 )
- 5 ml syringe
- 18 G needles (BD, catalog number: 305185 )
- 10 μl pipette tip
- Bottle top filter system (0.22 μm pore size)
- C57BL/6 male and females
- α-MEM (powder) (Gibco, catalog number: 11900-024 )
- Fetal Bovine Serum
Note: We used FBS from Atlanta Biological for this publication. However, we always perform lot check from 5-10 test FBS samples to find the best FBS for OP9 maintenance culture. We usually purchase 20 bottles to maintain the same equality of experiments for 2-3 years. - Fresh Milli-Q water
- PBS (Fisher Scientific, catalog number: BP665-1 )
- 2-Mercaptoethanol (Fisher Scientific, catalog number: O3446I-100 )
- 500 mM EDTA pH 8.0
- IMDM (Gibco, catalog number: 12440061 )
- Heparin (Sigma, catalog number: H3149-100KU )
- Endothelial mitogen 50 mg (Biomed Tech, catalog number: BT-203 )
- Penicillin-Streptomycin 10,000 U-10,000 µg/ml (Gibco, catalog number: 15140122 )
- X-VIVO 20 (Lonza, catalog number: 04-448Q )
- Methylcellulose-base medium for mouse pre-B lymphoid progenitor cells (Stemcell Technologies, MethocultTM, catalog number: M3630 )
- Recombinant murine IL-7 (Peprotech, catalog number: 217-17 )
- Recombinant murine Flt3-ligand (Peprotech, catalog number: 250-31L )
- 0.05% Trypsin/EDTA (Gibco, catalog number: 25300054 )
- 0.25% Collagenase Type I (Stemcell Technologies, catalog number: 0 7902 )
- DNase I Solution (1mg/ml) (Stemcell Technologies, catalog number: 0 7900 )
- Cell Dissociation Buffer (Hank’s balanced) (Gibco, catalog number: 13150-016 )
- Antibodies for sorting and detecting B cell colonies (Table 1)
Table 1. Antibody list for flow cytometry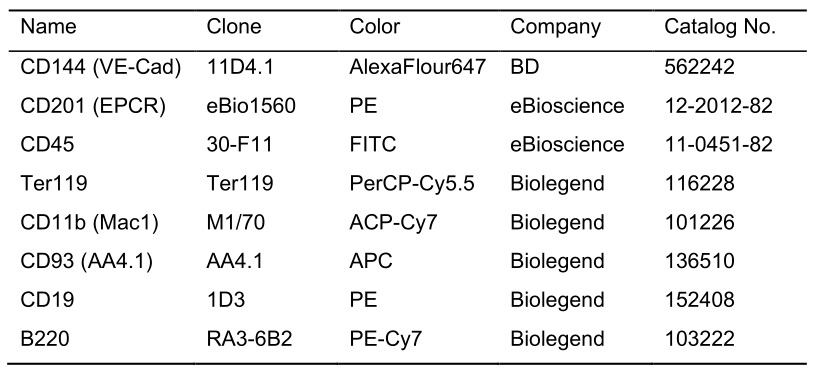
- OP9 stromal cells [obtained from Dr. Shin-Ichi Nishikawa (retired from Riken, Kobe, Japan), can be obtained from ATCC CRL-2749 (ideally < 30 passage)]
- Akt-expressing AGM-ECs (AGM-ECs): endothelial cells (ECs) obtained from aorta-gonod-mesonephros (AGM) region in the mouse embryo was established and induced Akt-overexpression by Dr. Brandon Hadland, Fred Hutchinson Cancer Center, Seattle, USA) (Hadland et al., 2015)
- Gelatin from porcine skin (Sigma, catalog number: G1890 )
- α-MEM (see Recipes)
- 2-mercaptoethanol (2-ME) stock (0.1 M) (see Recipes)
- OP9 maintenance medium (α-MEM + 20% FBS) (see Recipes)
- AGM-EC maintenance medium (see Recipes)
- Differentiation medium (α-MEM +10% FBS + 5 x 10-5 M 2ME) (see Recipes)
- Staining buffer (PBS + 5% FBS + 2 mM EDTA) (see Recipes)
- 0.1% Gelatin (see Recipes)
Equipment
- P1000 pipette
- Microcentrifuge
- FACS Sorter and analyzer with more than 6 fluorescent detectors are required. We used BD FACSAria for cell sorting, BD LSRII and BD LSR-Fortessa for analysis
- Standard cell culture CO2 incubator (CO2: 5%, O2: 20%)
Procedure
- Maintaining OP9 stromal cells and AGM-EC stromal cells
Maintaining OP9 stromal cells
- Maintaining OP9 stromal cells in a good condition is an important key to obtain successful results. Make fresh OP9 maintenance medium from the powder every month. Test several lots of FBS for good growth of OP9 cells.
- Thaw OP9 in T25 Flask in freshly made OP9 medium. Split every 3 days, 1:3.
Notes:
- OP9 cells are flat round or square shapes (Figure 1) if they become spindle shape, they are not good OP9 anymore because spindle shape cells tend to be transforming and losing B cell supporting ability.

Figure 1. The morphology of OP9 cells (100x). Red arrows indicate good OP9 and blue arrows indicate bad OP9. Scale bar = 100 μm. - Split one confluent OP9 in T25 flask into 3 flasks every 3 days. If they are not confluent in 3 days, the lot of FBS is not good for them.
- The density of OP9 needs to be appropriate (around 4 x 105/T25 flask at the starting point). When OP9 cells are confluent in T25 flask, the cell number is around 1.0 x 106-1.5 x 106.
- For splitting, aspirate medium, wash with PBS 2 times, and treat them with 0.05% Trypsin/EDTA for 3 min at 37 °C. OP9 cells will be peeled off from the flask and add medium and pipette to make a single cell suspension with 5 ml serological pipette (important). Make sure well pipetting and single cells, because leaving cell clumps may induce bad condition of OP9 cells at later passage. Spin 400 x g for 5 min, aspirate supernatant, loosen the pellets by tapping the bottom of the tube (important), add 1 ml OP9 maintenance medium and pipette very well with P1000 pipette to ensure single cell suspension. Add more OP9 medium and plate them into 3 x T25 flasks.
- OP9 must be maintained within 3 days (usually every 3 days). Even if they are not confluent in 3 days, they should be passed (1 to 1 or 1 to 2 to increase the cell density). If they are left confluent without passage, they can be used for co-culture, but they will not expand anymore.
- If OP9 cells are not in good condition, it is hard to obtain B cells from early stage of embryos (< E9.0-9.5).
- OP9 cells should not be maintained for more than 1 month.
- OP9 cells show good contact inhibition. If you need irradiation to stop the over-cell growth of OP9, they are already transformed and not good OP9 anymore.
- OP9 cells are flat round or square shapes (Figure 1) if they become spindle shape, they are not good OP9 anymore because spindle shape cells tend to be transforming and losing B cell supporting ability.
- Coat 0.1% gelatin T25 Flask for more than 30 min.
- Aspirate gelatin before plating cells.
- Thaw AGM-EC on gelatin-coated T25 Flask in AGM-EC maintenance medium (near 5 x 105/flask). Pass every 3-4 days, 1:1-3 (Hadland et al., 2015).
- Harvest embryo tissues
- Set up timed mating of C57BL/6 male and females after 3:00 PM. The following early morning, check vaginal plugs of the female and separate plugged females into a different cage. Noon on this day is considered as embryonic (E) day 0.5.
- About 7 days before harvesting embryos, thaw OP9 and AGM-ECs in T25 flask and maintain.
- One day before harvesting embryos, prepare OP9 (1 x 104/well) and AGM-ECs (5 x 103/well) in 96-well plates respectively. In addition, keep maintaining OP9 in T25 Flask as needed for plating methylcellulose at 5-7 days later.
- Ten days later after timed mating (at E10.5), harvest embryos from pregnant mother (follow the procedure under the approved AICUC protocol at your institute) (Morgan et al., 2008).
- Under the stereomicroscope, open the uterus, separate yolk sac (YS) from embryos (Morgan et al., 2008). After removal of the YS, confirm the embryonic stage by counting somite pairs. Collect caudal half of embryos, containing AGM region into 14 ml tubes filled with 10 ml medium.
- Centrifuge the AGM tissues in the medium at 450 x g for 5 min and aspirate supernatant. Suspend the AGM tissues in 0.25% Collagenase Type I + DNase I (final 20 μg/ml) respectively [use 0.5-1 ml per embryo equivalent (e.e)].
- Incubate the tissue suspension at 37 °C. Pipette every 5 min and confirm dissociation under microscope. Once you see good single cells, stop the collagenase reaction by adding the same volume of Cell Dissociation Buffer. Usually this incubation will take 15-30 min depending on the age of embryos (the earlier embryo, the shorter incubation time).
- Add differentiation medium and filter them using 70 μm strainer, and centrifuge the cells at 450 x g for 5 min.
- Aspirate supernatant, suspend single cell pellets in 1 ml staining buffer, and count cell number. Normally around 5 x 105 cells /embryo will be harvested.
- Sorting HECs/pre-HSCs and co-culture
When examining the B-lymphoid hematopoietic potential of HECs/pre-HSC, it is challenging to detect B-progenitor colony forming ability directly from these cells, since most of them are endothelial phenotype. In order to induce hematopoietic progenitors, sort HECs/pre-HSCs and plate them on OP9 (expand hematopoietic progenitors) or on AGM-ECs (let pre-HSCs mature into HSCs).
Staining and cell sorting- Suspend embryonic cells up to 1 x 107 cells in 100 μl staining buffer in 1.5 ml Eppendorf tube, take 5% volume first each for negative control and FMO staining.
- Add antibodies; anti-mouse CD45, CD144, c-kit, EPCR (CD201), and Ter119 at a ratio of 0.3 μl/106 cells and incubate them for 30 min on ice.
- Add 1 ml staining buffer for wash and centrifuge them at 800 x g for 2 min at microcentrifuge.
- Suspend the cells in staining buffer and sort Ter119-CD144+c-kit+EPCR+ cells on FACSAria (Figure 2).
- (optional) mix sorted Ter119-CD144+c-kit+EPCR+ cells with 1 x 105 OP9 cells in Methocult with IL-7 and Flt3 -ligand, and plate them onto a 35 mm Petri dish. However, direct plating of these cells will yield only a few B cell colonies.
- Plate 100-200 Ter119-CD144+c-kit+EPCR+ cells at one well of 96-well plate confluent with OP9 in differentiation medium or AGM-EC cells in X-vivo 20 with added IL-7 and Flt3-ligand (final concentration: 10 ng/ml for both).
- Five to seven days after co-culture, harvest all cells and stain them with anti-mouse CD45 and CD11b antibody.
- After staining, cells are washed with staining buffer and are suspended in staining buffer and sort CD11b-CD45+ cells from the co-cultured cells on FACSAria as shown in Figure 2B.
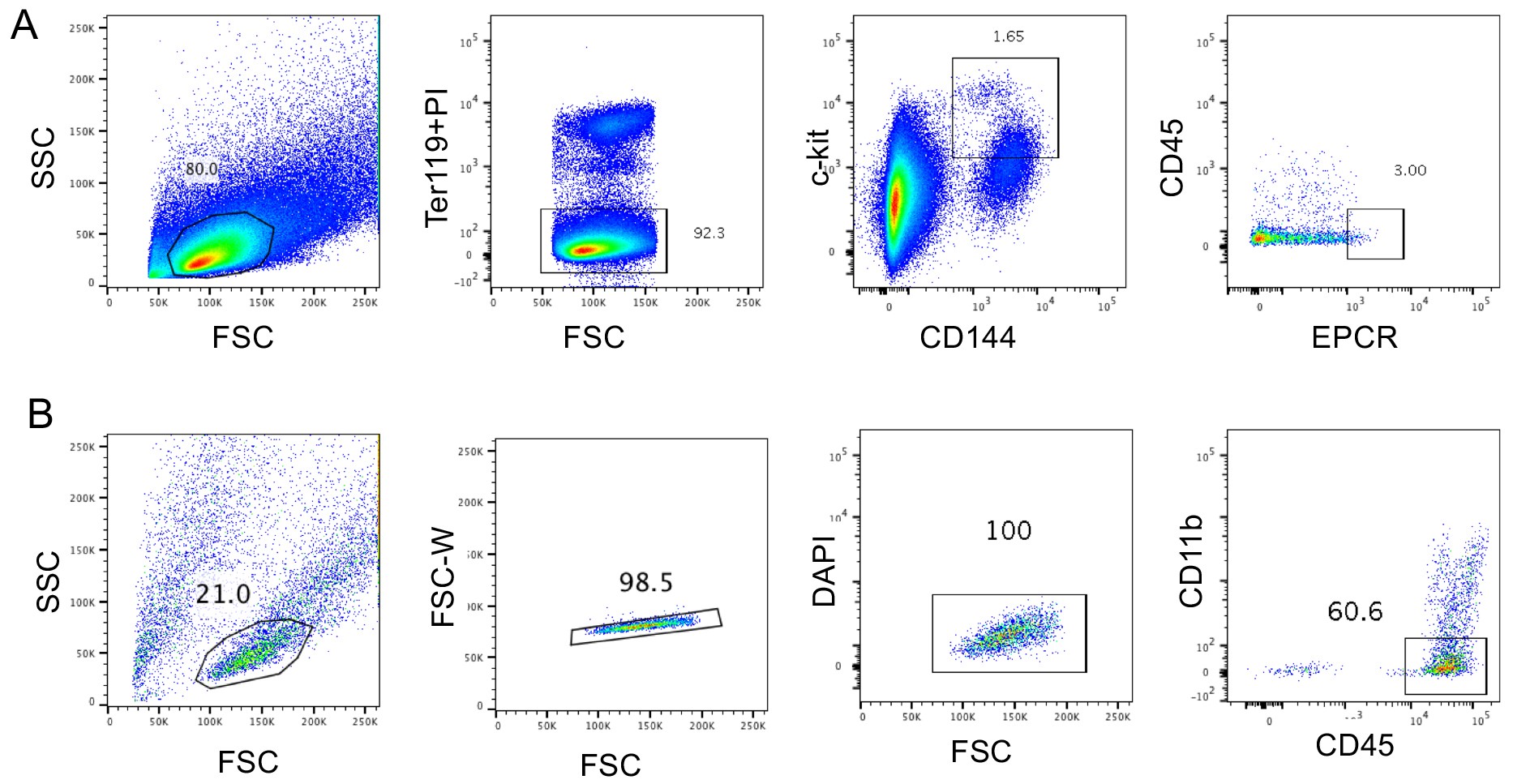
Figure 2. Gating strategy for sorting pre-HSCs (A) and CD45+CD11b- cells after co-culture with AGM-ECs(B)
- Plating semisolid culture
- Thaw Methocult at room temperature or 4 °C over night.
- Always prepare N+1 samples for methylcellulose assays. Mix 200-600 CD45+ cells after co-culture with 1 x 105 OP9 cells/dish. For triplicates, mix 800-2,400 CD45+ cells and 4 x 105 OP9 cells in 14 ml round bottom tube and spin them down, aspirate the medium, and loosen the cell pellets by tapping the tube. Add 4 ml Methocult M3630 using 5 ml syringe with 18 G needle to the cell pellet (This way, you can plate 200-600 CD45+ cells with 1 x 105 OP9 cells per 35 mm dish.). In this scale, 20-80 colonies/dish will be expected.
- Add IL-7 (final 10 ng/ml) and Flit3-ligand (final 10 ng/ml), mix well by vortexing and leave it until all the bubbles are gone.
- Plate 1.1 ml of methylcellulose medium including cells into each 35 mm Petri dish x 3 dishes using 18 G needle with 3 ml syringe.
- Place 3 Petri dishes containing the Methocult and 1 Petri dish containing sterile H2O (preventing the dry out the medium) in a 15 mm Petri dish.
- Incubate them in a 5% CO2 incubator for 8-11 days and count colony numbers. The pictures in Figure 3 are representative B-cell colonies and their FACS analysis.
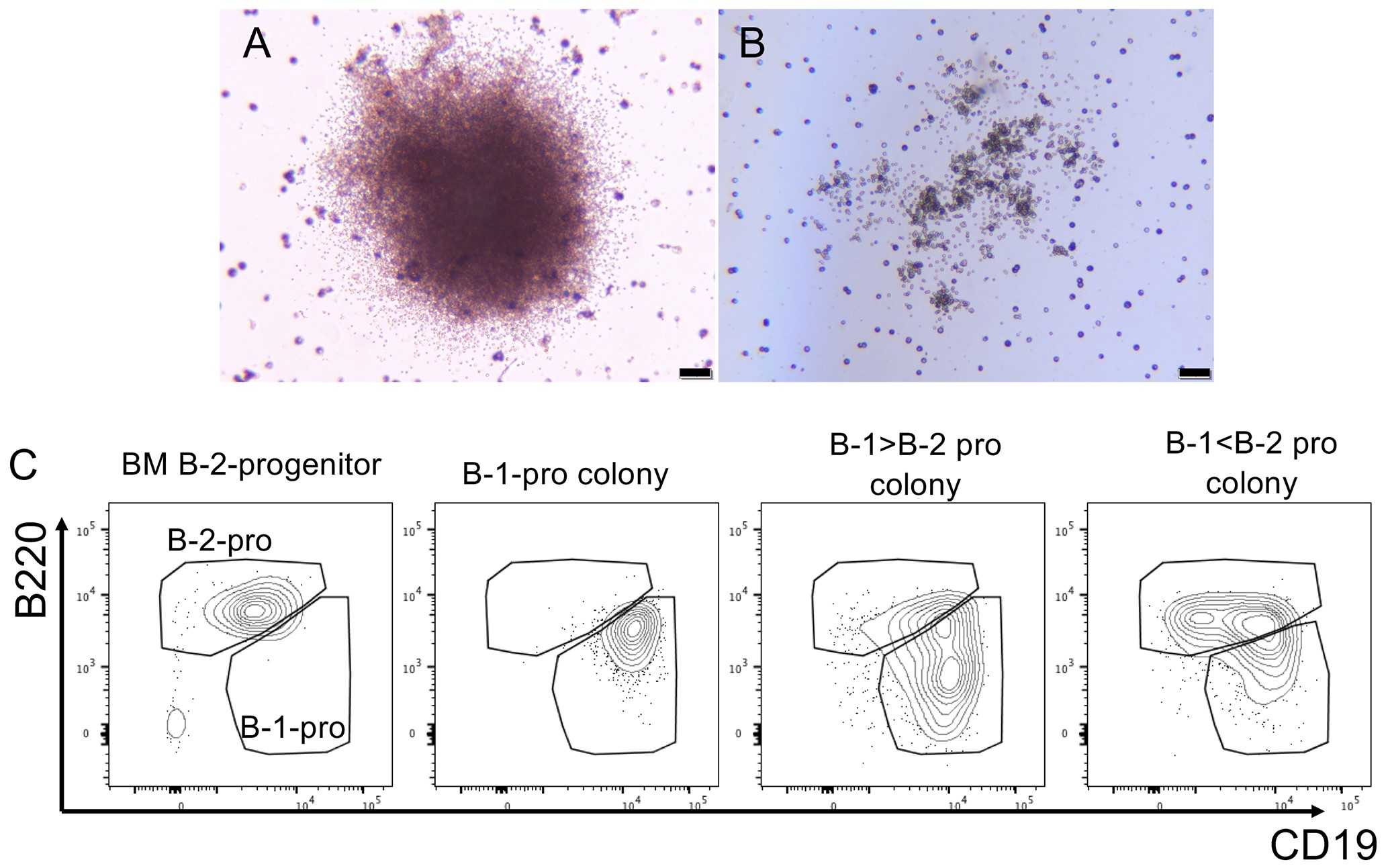
Figure 3. B cell colonies and FACS analysis of picked-up colonies. A. B-1 progenitor (B-1-pro) colony. B. B-2 progenitor (B-2-pro) colony, confirmed by FACS analysis below. Scale bar = 200μm. C. FACS analysis of each picked up colony. Based on the gating of BM B-2 progenitors, same gating is applied to the analysis of each sample.
- Determine each colony as to B-1 or B-2 progenitors
- Prepare 10 μl pipette and 1 ml differentiation medium in sterile Eppendorf tubes. For picked up colonies, prepare 1.5 ml Eppendorf tubes containing 1 ml staining buffer. Prepare the same number of tubes as the colony number.
- Before picking up the colony, wet the 10 μl pipette tip with differentiation medium by pipetting several times to avoid the methylcellulose sticking to the inside of the pipette tips. Set 8 μl to pick up a colony. Under the microscope, pick up each colony using 10 μl pipette with a wet tip, and transfer each colony to the 1.5 ml tube containing staining buffer.
- Once all the colonies are picked up into each tube, centrifuge the tubes at 800 x g for 2 min in the microcentrifuge and aspirate the supernatant.
- Add 50 μl of staining buffer and vortex to make the pellet a single cell suspension.
- Stain them with the antibodies: anti-mouse AA4.1-APC, CD19-PE, B220-PEcy7, and Mac1-FITC at a ratio of 0.3 μl/106 cells for 20-30 min on ice.
- Wash with 1 ml staining buffer and centrifuge at 800 x g for 2 min.
- Aspirate the supernatant and suspend the cell pellet with staining buffer with PI or DAPI for dead cell exclusion and apply the samples to flow cytometry.
- Determine whether the colonies contain B-1 and/or B-2 progenitors (Figure 3) (Kobayashi et al., 2019). Prepare adult BM cells as a positive control for gating B-progenitors (Montecino-Rodriguez et al., 2006).
Data analysis
Data analysis and expected outcome
Based on the flow cytometric analysis, each dish should contain 1) only B-1 progenitors, 2) only B-2 progenitors, and 3) both B-1 and B-2 progenitors. Count number of colonies that contain B-1, B-2, or both B-1 and B-2 progenitors per plate and calculate mean and SD values for the results of three dishes.
Usually, only B-2 colonies are expected from BM progenitors. B-1 progenitor colonies are dominant from embryonic tissues co-cultured with OP9, while B-1+B-2 progenitor colonies become dominant when these embryonic tissues are co-cultured with AGM-ECs or organ cultured (developing to HSCs) (Kobayashi et al., 2019).
Notes
- AGM-ECs can support the transition of pre-HSCs to HSCs, but they do not support B cell colony formation when they are plated together in Methylcellulose culture.
- The number of B cell colony forming cells varies depending on the age of the embryo and the condition of the OP9.
- Ter119-CD144+c-kit+EPCR+ cells are very rare, therefore, Ter119-CD144+c-kit+ should also be able to produce B cell colonies by support of OP9.
- Do not add SCF in the co-culture nor methylcellulose culture. Good OP9 support B-lymphopoiesis without SCF and SCF may enhance macrophage differentiation.
Recipes
Culture medium:
- α-MEM (with penicillin/streptomycin 50 U-50 μg/ml)
- Add powder α-MEM to 1,000 ml fresh Milli-Q water and stir until it is completely dissolved
- Add 2.2 g H2CO3 and stir until completely dissolved
- Add 5 ml Penicillin-Streptomycin
- Filter using 0.22 μm filtering system
- 2-mercaptoethanol (2-ME) stock (0.1 M)
Mix 70 μl of 2-ME to 10 ml α-MEM and filter using 0.22 μm syringe filter - OP9 maintenance medium (α-MEM + 20% FBS)
Mix 400 ml α-MEM and 100 ml FBS and filter using 0.22 μm filtering system - AGM-EC maintenance medium
IMDM 400 ml
FBS (Hyclone, heat inactivate)100 ml
Pen/Strep (100x) 5 ml
Heparin 5 ml (10 mg/ml stock prepared fresh in IMDM)
L-glutamine (100x) 5 ml
Endothelial mitogen 50 mg - Dissolve in 10 ml of above media mix
- Filter using 0.22 μm filtering system
- Differentiation medium (α-MEM + 10% FBS + 5 x 10-5 M 2-ME)
450 ml α-MEM
50 ml FBS
250 μl of 0.1 M 2-ME
Filter using 0.22 μm filtering system - Staining buffer (PBS + 5% FBS + 2 mM EDTA)
500 ml PBS 500 ml
2.5 ml FBS + EDTA (final 2 mM)
Filter using 0.22 μm filtering system - 0.1% Gelatin
0.5 g Gelatin
Milli-Q water 500 ml
Autoclave
Acknowledgments
This work is supported by NIAID R01AI121197. This protocol was derived from a recent report (Kobayashi et al., 2019).
Competing interests
There is no conflict of interest.
Ethics
The procedure of harvesting mouse embryos and other related animal works follows the Animal Welfare Committee (AWC) protocol approved by the Center for Laboratory Animal Medicine and Care (CLAMC) at UTHealth (AWC16-0124, 2016-2019).
References
- Hadland, B. K., Varnum-Finney, B., Poulos, M. G., Moon, R. T., Butler, J. M., Rafii, S. and Bernstein, I. D. (2015). Endothelium and NOTCH specify and amplify aorta-gonad-mesonephros-derived hematopoietic stem cells. J Clin Invest 125(5): 2032-2045.
- Kobayashi, M., Tarnawsky, S. P., Wei, H., Mishra, A., Azevedo Portilho, N., Wenzel, P., Davis, B., Wu, J., Hadland, B. and Yoshimoto, M. (2019). Hemogenic Endothelial Cells Can Transition to Hematopoietic Stem Cells through a B-1 Lymphocyte-Biased State during Maturation in the Mouse Embryo. Stem Cell Reports 13(1): 21-30.
- Montecino-Rodriguez, E., Fice, M., Casero, D., Berent-Maoz, B., Barber, C. L. and Dorshkind, K. (2016). Distinct Genetic Networks Orchestrate the Emergence of Specific Waves of Fetal and Adult B-1 and B-2 Development. Immunity 45(3): 527-539.
- Montecino-Rodriguez, E., Leathers, H. and Dorshkind, K. (2006). Identification of a B-1 B cell-specified progenitor. Nat Immunol 7(3): 293-301.
- Morgan, K., Kharas, M., Dzierzak, E. and Gilliland, D. G. (2008). Isolation of early hematopoietic stem cells from murine yolk sac and AGM. J Vis Exp(16): pii: 789.
- Yoshimoto, M., Montecino-Rodriguez, E., Ferkowicz, M. J., Porayette, P., Shelley, W. C., Conway, S. J., Dorshkind, K. and Yoder, M. C. (2011). Embryonic day 9 yolk sac and intra-embryonic hemogenic endothelium independently generate a B-1 and marginal zone progenitor lacking B-2 potential. Proc Natl Acad Sci U S A 108(4): 1468-1473.
Article Information
Copyright
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Kobayashi, M. and Yoshimoto, M. (2020). A Modified Semisolid Clonal Culture for Identification of B-1 and B-2 Progenitor Colony Forming Ability of Mouse Embryonic Hemogenic Endothelial Cells. Bio-protocol 10(9): e3601. DOI: 10.21769/BioProtoc.3601.
Category
Developmental Biology > Cell growth and fate > Differentiation
Stem Cell > Embryonic stem cell > Cell-based analysis
Cell Biology > Cell isolation and culture > Co-culture
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link