- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Enzymatic Construction of Protein Polymer/Polyprotein Using OaAEP1 and TEV Protease
Published: Vol 10, Iss 8, Apr 20, 2020 DOI: 10.21769/BioProtoc.3596 Views: 4880
Reviewed by: Beatrice LiHongwei HanAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Production, Titration and Imaging of Zika Virus in Mammalian Cells
Wesley Freppel [...] Laurent Chatel-Chaix
Dec 20, 2018 11814 Views

Purification and Proteomic Analysis of Alphavirus Particles from Sindbis Virus Grown in Mammalian and Insect Cells
Raquel Hernandez [...] Davis F. Ferreira
May 20, 2019 7507 Views

Identification of Buffer Conditions for Optimal Thermostability and Solubility of Herpesviral Protein UL37 Using the Thermofluor Assay
Andrea L. Koenigsberg [...] Ekaterina E. Heldwein
Jun 20, 2020 3873 Views
Abstract
The development of chemical and biological coupling technologies in recent years has made possible of protein polymers engineering. We have developed an enzymatic method for building polyproteins using a protein ligase OaAEP1 (asparagine endopeptidase 1) and protease TEV (tobacco etching virus). Using a mobile TEV protease site compatible with the OaAEP1 ligation, we achieved a stepwise polymerization of the protein on the surface. The produced polyprotein can be verified by protein unfolding scenario using atomic force microscopy-based single-molecule force spectroscopy (AFM-SMFS). Thus, this study provides an alternative method for polyprotein engineering and immobilization.
Keywords: PolymerizationBackground
Several methods based on biochemical reactions for protein polymerization have been developed. For example, one approach is to design protein monomer with additional cysteines as the basic unit for ligation (Dietz et al., 2006; Durner et al., 2017). Another approach is to build the complete gene for the artificial protein oligomer (Carrion-Vazquez et al., 1999; Giganti et al., 2018). However, the engineering of large-sized protein polymer is often challenging. To address this challenge, we have developed an enzyme-mediated method that builds protein polymers/polyproteins in a stepwise fashion, using protein ligase and protease (Yang et al., 2017; Deng et al., 2019). When a TEV site (ENLYFQ/G) plus a leucine (L) is designed as ENLYFQ/GL-POI (POI: Protein of Interest) at the N-terminus of the protein unit, TEV cleavage produces an N-terminal GL residue of GL-POI, which is compatible with further OaAEP1 ligation. Our enzymatic method provides a new method for the preparation of the polyprotein sample with a controlled sequence and suitable for single-molecule studies, especially for complex protein systems.
The engineered polyprotein sequence is confirmed by individual protein unfolding event using AFM-SMFS. Besides a powerful imaging tool (Mannix et al., 2018), AFM can manipulate single molecule mechanically and directly measure its unfolding, unbinding, and rupture force (He et al., 2012; Scholl and Marszalek, 2018; Zhang et al., 2019). Thus, it is widely used to study protein (un)folding and mechanics (Infante et al., 2019; Krieg et al., 2019), protein-protein/ligand-receptor interaction (Ott et al., 2017; Zhang et al., 2019) and even chemical bond (Pill et al., 2019; Song et al., 2019; Yuan et al., 2019). Together with other established polyprotein engineering and immobilization methods (Dietz et al., 2006; Popa et al., 2013; Hoffmann et al., 2015; Walder et al., 2017), our enzymatic methodology can improve the quality and efficiency of SMFS study.
Materials and Reagents
- Glass coverslip (Sail Brand, China)
- E. coli BL21(DE3) (-80 °C)
- pQE80L-POI or pET28a-POI plasmid (Vector information in the Supplemetal file)
- Milli-Q water (18.2 MΩ/cm)
- Luria-Bertani (LB) medium (BD Difco)
- Iron (III) chloride hexahydrate (99%, Energy chemical)
- Calcium chloride hydrate crystalline aggregate (99.9965%, Alfa Aesar)
- (3-aminopropyl) triethoxysilane (APTES, 99%, Sigma-Aldrich)
- Sulfosuccinimidyl 4-(N-maleimidomethyl) cyclohexane-1-carboxylate (sulfo-SMCC, Thermo Scientific)
- Sodium chloride (NaCl) (99%, BBL Life Science)
- Tris(hydroxymethyl) aminomethane (Tris, > 99.9%, Sangon Biotech)
- EDTA (99.5%, Macklin)
- 5,5'-Dithiobis (2-nitrobenzoic Acid) (99%, Alfa Aesar)
- Ampicillin sodium salt (99% Zhu YanBIO)
- Kanamycin (≥ 750 μg/mg, Diamond)
- Magnesium sulfate (MgSO4) (AR, Shanghai Lingfeng Chemical Reagent Co., Ltd)
- Calcium chloride (CaCl2) (AR, Aladdin)
- D(+)-Glucose (AR, Aladdin)
- IPTG (Isopropyl-β-d-thiogalactopyranoside) (99% Zhu YanBIO)
- DNase (from Bovine Pancreas, Activity ≥ 500 Kunitz U/mg, Sangon Biotech)
- RNase (from Bovine Pancreas, Activity ≥ 60 U/mg, Sangon Biotech)
- PMSF (Zhu YanBIO)
- Potassium chromate (99.5%, Macklin)
- Concentrated sulfuric acid (AR, Sinopharm Chemical Reagent Co.,Ltd)
- Ethyl alcohol (AR, Sinopharm Chemical Reagent Co.,Ltd)
- Imidazole (99% Aladdin)
- Urea (AR, Sinopharm Chemical Reagent Co.,Ltd)
- TEV protease (produced by ourselves)
- OaAEP1 (produced by ourselves)
- BamHI (TaKaRa)
- BglII (TaKaRa)
- KpnI (TaKaRa)
- T4 Ligase (TaKaRa)
- Glycerol (99%, Macklin)
- M9 medium (see Recipes)
- Chromic acid (see Recipes)
- Lysis buffer (see Recipes)
- Wash buffer (see Recipes)
- Elution buffer (see Recipes)
- DTNB solution (see Recipes)
- TEV protease solution (see Recipes)
- AFM measurement buffer (see Recipes)
Equipment
- Avanti JXN-30 Centrifuge (Beckman Coulter)
- AKTA FPLC system (GE Healthcare)
- Mono Q 5/50 GL (GE Healthcare)
- NanoDrop 2000 spectrophotometer (Thermo Scientific)
- Nanowizard 4 AFM (JPK Instruments AG)
- Silicon nitride cantilever (MLCT, Bruker Corp)
- Sonictor (Biosafer 650-92)
Software
- JPK data processing (developed by JPK Instruments AG)
- Igor Pro 6.12 (Wavemetrics)
Procedure
- Gene cloning
- Use three-restriction digestion enzyme system BamHI-POI-BglII-stop codon-KpnI for connecting the gene of different protein fragments. In this enzyme system use BglII and KpnI digest to produce vector and use BamHI and KpnI digest to produce the insert. As BamHI digestion and BglII digestion leads to the same cohesive end (GATC), it is possible to use T4 ligase connect the vector and the insert.
- Confirm all genes by sequencing.
- Expression and production of interest proteins
Note: In the experimental operation, always pay attention to the aseptic operation. Different proteins have different E. coli harvest shelf life. It is necessary to test the shelf life for the specific protein.- Transform the pQE80L-POI or pET28a-POI plasmid into E. coli BL21(DE3) cell.
- Apply the bacterial solution to the LB plate containing ampicillin sodium salt (100 μg/ml) for pQE80L plasmid or kanamycin (50 μg/ml) for pET28a plasmid and incubate at 37 °C for 14-16 h.
- Pick single colonies into 15 ml LB medium containing ampicillin sodium salt (100 μg/ml) or kanamycin (50 μg/ml). Keep shaking at 200 rpm at 37 °C for 16-20 h.
- After grown saturation, dilute the overnight cultures into 800 ml fresh LB media (add to the LB medium in a ratio of 1:50) containing respective antibiotics (the concentration of antibiotics is the same as above). As for the metalloprotein, centrifuged the culture at 1,800 x g and resuspended the precipitation by 16 ml M9 medium, and then add to 800 ml respective antibiotics containing M9 medium as well.
- Shake culture at 200 rpm at 37 °C until the optical density at 600 nm (OD600) reaches 0.6-0.8 (this takes about 2.5-3 h, while M9 medium always takes about 5-6 h). Reserve a 100-μl sample of the culture as the pre-induction control for testing protein expression.
- Add IPTG (Isopropyl-β-d-thiogalactopyranoside) to a final concentration of 1 mM to induce protein expression, while metalloprotein always needs some additional metal ion, and shake the culture at 200 rpm at 37 °C for 4-6 h.
- Harvest the culture at 13,000 x g for 25 min at 4 °C and store at -80 °C before purification.
- Purification of interest proteins
- Resuspend the cells in 25 ml lysis buffer (contained 0.06 mg DNase, 0.06 mg RNase, 0.08 mg PMSF) and produce the lysis using a Biosafer sonicator (15% amplitude) on ice for 30 min. Clarify the cell lysate at 19,600 x g for 40 min at 4 °C and collect the supernatant fraction.
- Pack a Poly-Prep column with 1-1.5 ml (bed volume) of Co-NTA or Ni-NTA affinity column and wash it with ten column volumes (CV) of Milli-Q water and then 10 CVs wash buffer (by gravity flow).
- Load the supernatant onto the column by gravity flow and collect the flow-through. Load the flow-through onto the column by gravity and collect the secondary flow-through. Load the secondary flow-through onto the column by gravity and finish the sample loading.
- Wash the column with 50 CVs of wash buffer.
- Elute the bound protein with 3 CVs of elution buffer at 4 °C for 15 min. Moreover, carry out anion exchange purification for pure metal form rubredoxin protein using AKTA with Mono Q column at pH 8.5 at 4 °C.
- Analyze the samples of the eluate and control group by SDS-PAGE.
- Use the Ellman method to confirm the concentration of ELPs, in which 10 μl protein incubated with 20 μl DTNB solution for 30 min at room temperature and the solution composited with 10 μl the elution buffer solution and 20 μl DTNB solution served as blank to detect the absorbance of protein and DTNB reaction solution at 412 nm (ε = 13,700 M−1 cm−1) by NanoDrop 2000 spectrophotometer.
- Functionalized coverslip surface preparation
Note: The functionalized coverslip should be immediately used or stored at 4 °C for a month after preparation.- Clean and activate glass coverslips by immersing in chromic acid at 80 °C for 30 min. Wash away chromic acid traces on the coverslips by water and then ethyl alcohol. Dry the coverslips by a stream of nitrogen.
- Aminosilylate the coverslips by immersing them in 1% (v/v) APTES toluene solution for 1 h at room temperature avoiding light.
- Wash the coverslips with toluene and absolute ethyl alcohol and dry the coverslips with nitrogen.
- Bake the coverslips at 80 °C for 15 min.
- Add 200 μl sulfo-SMCC (1 mg/ml) dimethyl sulfoxide (DMSO) solution between two immobilized coverslips after the coverslips cooling down to room temperature and incubate for 1 h protected from light at room temperature.
- Wash every piece of the coverslips with 15 ml DMSO first and then with absolute ethyl alcohol to remove residual sulfo-SMCC. Use a stream of nitrogen to dry the coverslips.
- Stick cleaned quartz ring on the functioned side of the coverslip to build a chamber.
- Pipet 200 μl of 200 μM GL-ELP50nm-C protein solution onto the functionalized coverslip in the chamber and incubate for approximately 3 h.
- Wash the chamber with Milli-Q water to remove the unreacted GL-ELP50nm-C.
- Functionalized cantilevers surface preparation
Note: Silicon nitride cantilever was used as a force probe. The surface chemistry of the cantilevers was similar to that of the coverslip.- Immerse the cantilevers at 80 °C for 10 min by chromic acid treatment. Remove the traces of chromic acid by water first and then ethyl alcohol. Use a piece of filter paper blot ethyl alcohol traces up.
- Functionalize the cleaned cantilevers by amino-silanization with APTES and then conjugate the cantilevers to sulfo-SMCC as the method mentioned in Section D, Functionalized coverslip surface preparation.
- Immerse the functionalized cantilever with Cys-ELP50nm-NGL at the concentration of 200 μM to the surface with maleimide group of sulfo-SMCC for 1.5 h.
- Wash the coverslip with Milli-Q water to remove the unreacted Cys-ELP50nm-NGL and store the cantilever at -20 °C.
- Before the AFM experiment, immerse a ELP-functionalized cantilever in 50 μM GL-CBM-XDoc protein solution with 200 nM OaAEP1 for 20-30 min at 25 °C, and then wash away the unreacted proteins by 15-20 ml AMF buffer.
- Stepwise polyprotein preparation with controlled sequences
Note: Use 15-20 ml AFM buffer to wash away residual proteins after each reaction step. Figure 1 presents the schematic diagram of the controlled sequence polyprotein preparation.- Mix unit Coh-tev-L-POI-NGL(50 μM) and OaAEP1 (200 nM) and add 60 μl to the functionaliazed coverslip. link the ligation unit Coh-tev-L-POI-NGL to the functionalized coverslip.
- Add 100 μl TEV protease solution to cleave the TEV site in the protein unit for about 1 h at 25 °C. And GL-Ub-NGL-glass will produce with exposed N-terminal GL residues.
- Repeat Steps F1-F2 (N-1) times to get the protein-polymer GL-(Ub)N-NGL-Glass. After OaAEP ligation of the final ligation unit, do not take the TEV cleavage reaction to obtain Coh-tev-L-(Ub)n-NGL-Glass.
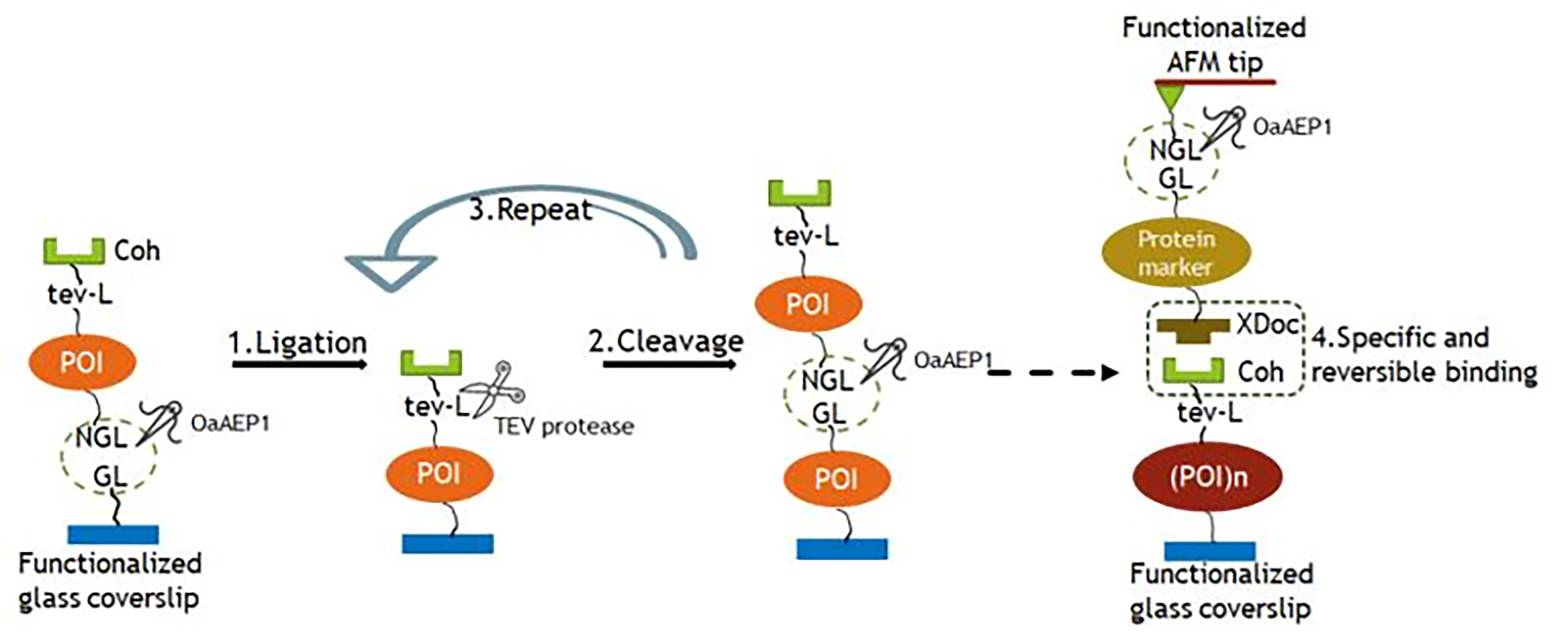
Figure 1. The schematic displays the stepwise ligation and cleavage procedure to produce sequence-controlled polyprotein on the glass surface and probed with an XDoc-protein maker-modified cantilever
- AFM measurements
Note: The AFM experiment parameters (Z length, Z speed, setpoint, sample rate and duration time) are adjustable in different experiments.- Add 1 ml AFM buffer containing10 mM CaCl2 to the chamber (Figure 2).
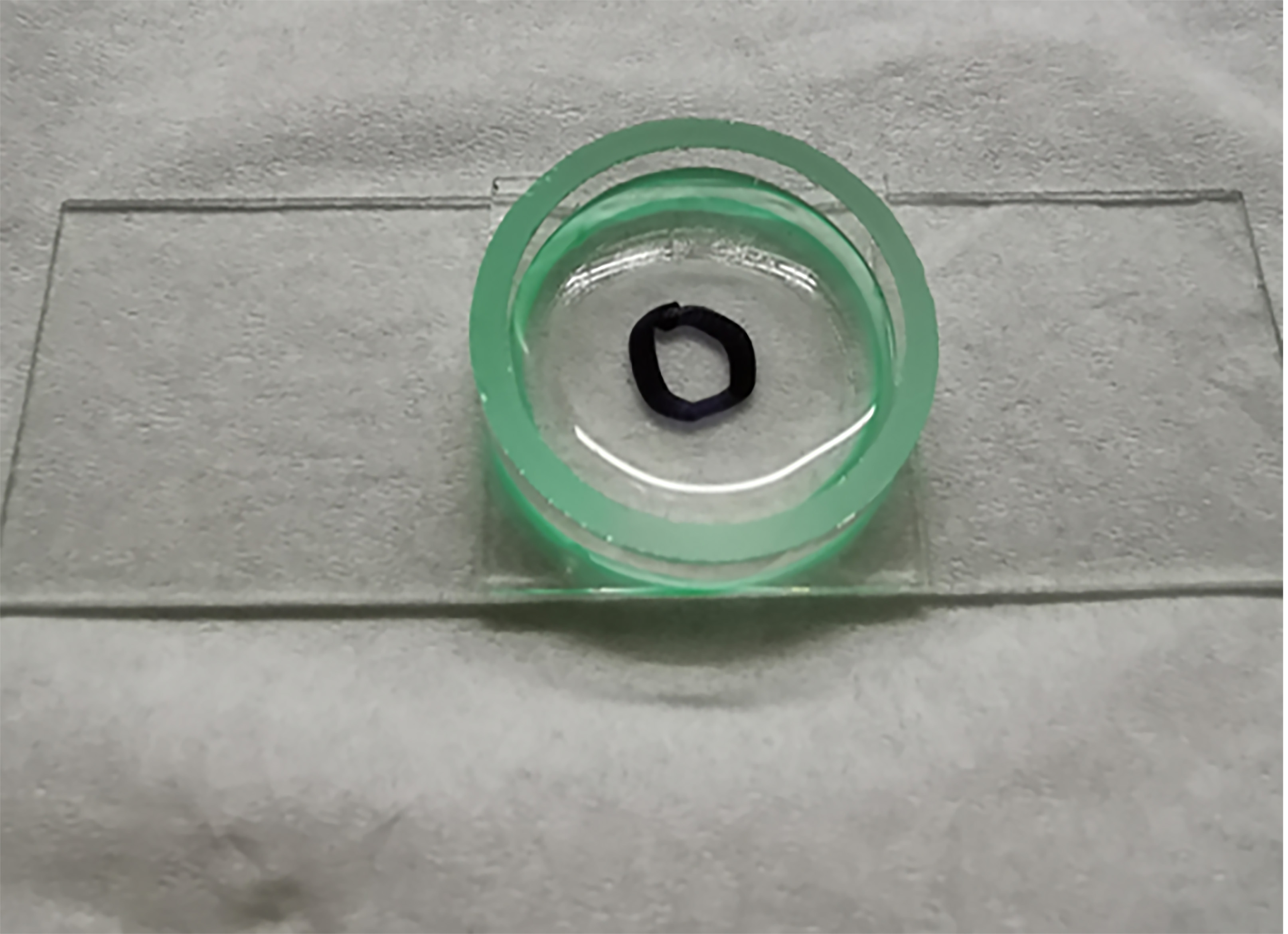
Figure 2. A photo image about AFM chamber with 1 ml AFM buffer solution. The black ring shows the position of the polyprotein. - Move the laser to the D tip of the functionalized probe (Figure 3). Use the equipartition theorem to calibrate the spring constant (κ) of each cantilever in solution with an accurate value before the experiment.
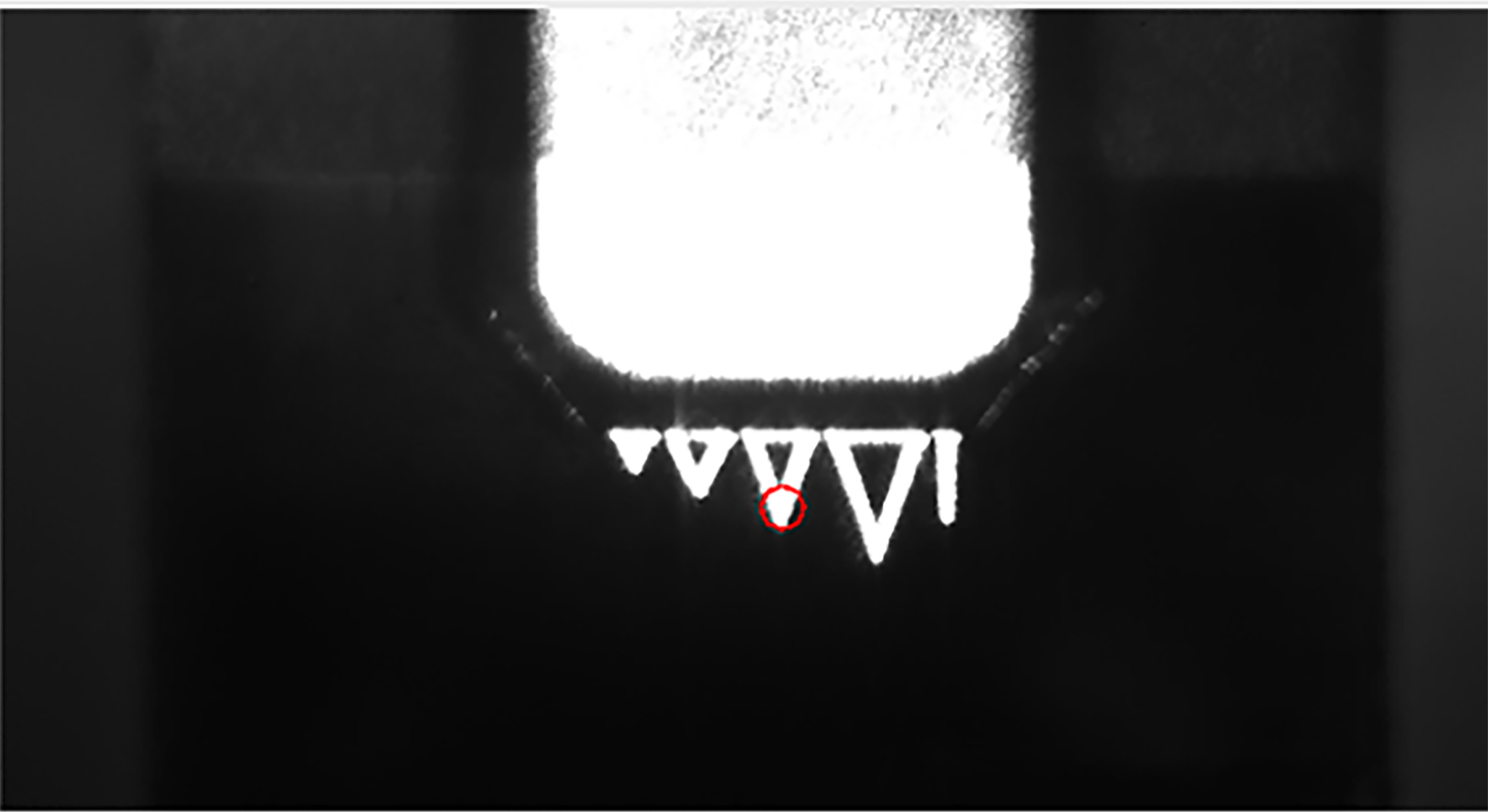
Figure 3. CCD picture of the cantilever tip in the AFM experiment. The triangle which the red ring located is D tip. The red ring points out the lase radiation position on D tip. - The cantilever tip attached to the sample surface and hold at this position for a while to formed the cohesin/dockerin pair as shown in Figure 4, segment 0 and segment 1.
- Retracted the cantilever from the surface at a constant velocity of 400 nm s−1 while recording the distance and cantilever deflection at a sampling rate of 4,096 Hz. As shown in Figure 4, segment 2.
- Hold the cantilever for a while to relax the cohesin/dockerin pair as shown in Figure 4, segment 3.
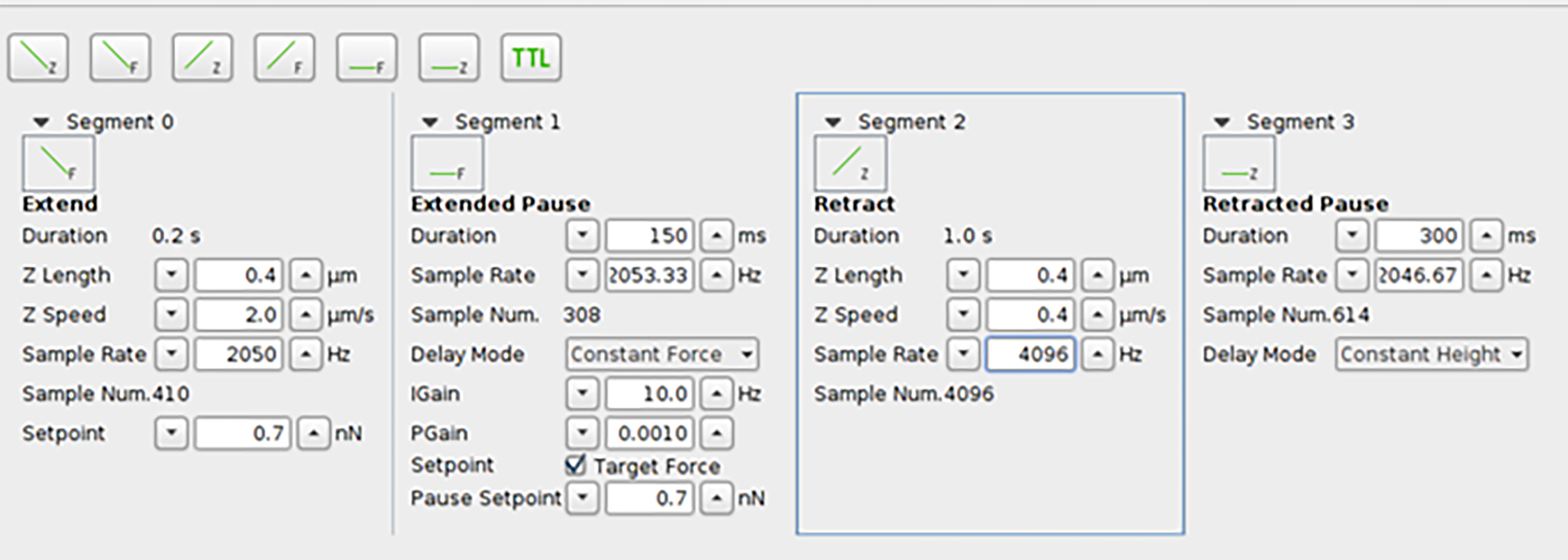
Figure 4. The dialog about the AFM experiment parameter setting. One AFM measurement contain four step. Segment 0 makes cantilever move towards the sample surface. Segment 1 keep the cantilever staying at the height reached by segment 0 for duration time. Segment 2 retract the cantilever from the surface and the protein unfold during this period. Segment 3 keep the cantilever at the height reached by segment 2 for duration time.
- Add 1 ml AFM buffer containing10 mM CaCl2 to the chamber (Figure 2).
Data analysis
- Use JPK data processing select force-extension traces.
- Use Igor Pro 6.12 (Wavemetrics) analysis the data traces. Only the data traces containing all the specific contour length increments belong to fingerprint protein domains [GB1(H18 nm) and CBM (H58 nm)] and a high rupture force from the cohesin-dockerin dissociation meet the requirement (more than 200-300 pN) (Figure 5). Fit The curves by the worm-like-chain (WLC) model of polymer elasticity with the persistence length of ~0.4 nm.
- Use Gauss model fit the histograms of unfolding forces to determine values of the most probable unfolding force (
) and contour length increment (<∆Lc>). 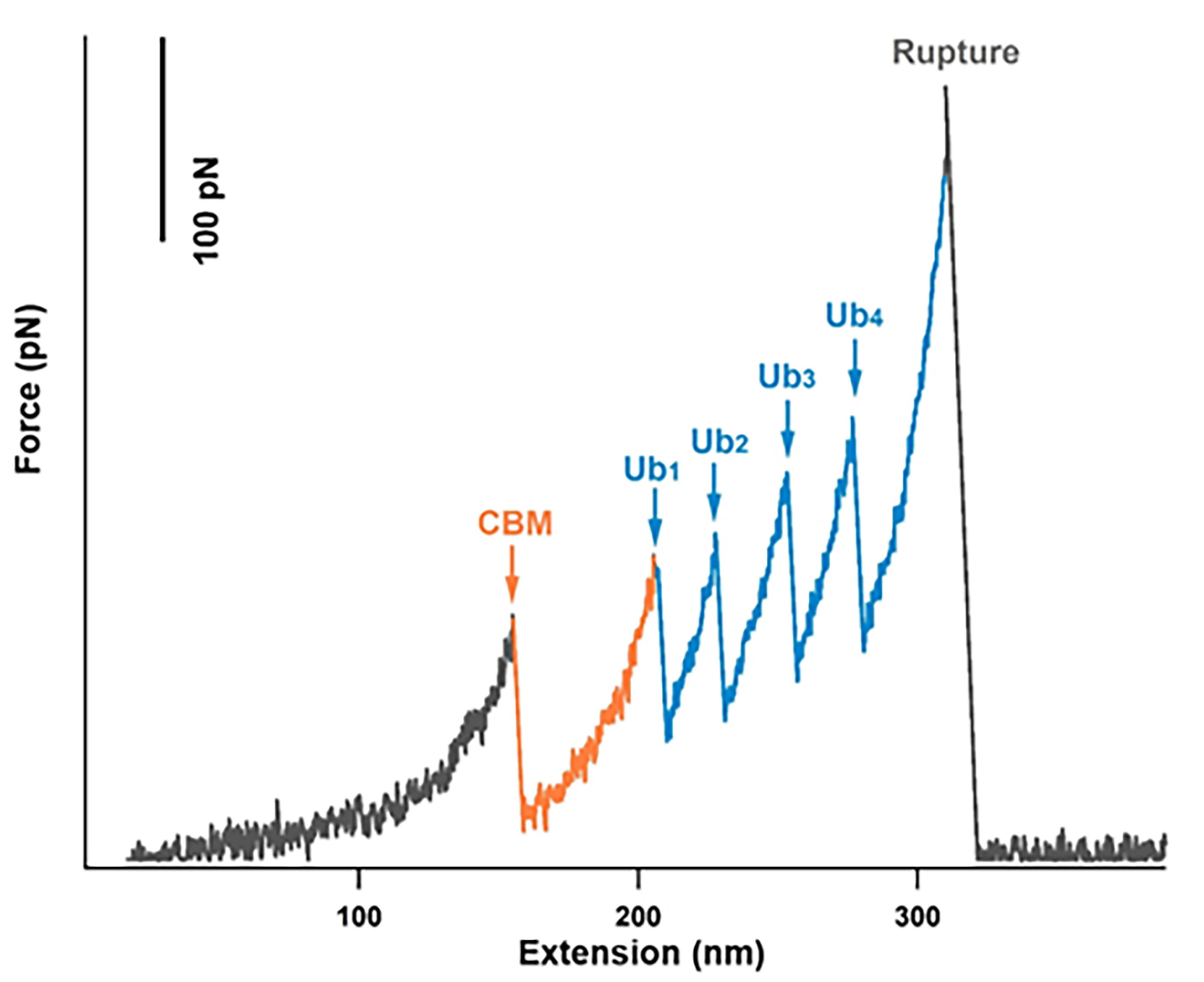
Figure 5. Typical force versus distance trace for the sequence-controlled polyprotein, Coh-Ub4, containing signals of CBM unfolding, dockerin-cohesin pair rupture and four copies of Ub
Recipes
- Lysis buffer
50 mM Tris
150 mM NaCl
pH 7.4 - Wash buffer
20 mM Tris
400 mM NaCl
2 mM imidazole
pH 7.4 - Elution buffer
20 mM Tris
400 mM NaCl
250 mM imidazole
pH 7.4 - DTNB solution
0.5 mM 5,5'-Dithiobis (2-nitrobenzoic Acid)
1 mM EDTA
7.2 M Urea - TEV protease solution
0.4 mg/ml TEV protease
75 mM NaCl
0.5 mM EDTA
25 mM Tris-HCl, pH 8.0
10% [v/v] glycerol - AFM measurement buffer
100 mM Tris
100 mM NaCl
pH 7.4 - Chromic acid
20 g potassium chromate
40 ml ultrapure water
360 ml concentrated sulfuric acid - M9 medium
0.4% glucose
0.1 mM CaCl2
2 mM MgSO4
Acknowledgments
National Natural Science Foundation of China (Grant Nos. 21771103, 21977047), Natural Science Foundation of Jiangsu Province (Grant No. BK20160639). This protocol was modified from previous work “Enzymatic biosynthesis and immobilization of polyprotein verified at the single-molecule level (Deng et al., 2019).
Competing interests
The authors declare no conflict of interest.
References
- Carrion-Vazquez, M., Oberhauser, A. F., Fowler, S. B., Marszalek, P. E., Broedel, S. E., Clarke, J. and Fernandez, J. M. (1999). Mechanical and chemical unfolding of a single protein: A comparison. Proc Natl Acad Sci U S A 96(7): 3694-3699.
- Deng, Y., Wu, T., Wang, M., Shi, S., Yuan, G., Li, X., Chong, H., Wu, B. and Zheng, P. (2019). Enzymatic biosynthesis and immobilization of polyprotein verified at the single-molecule level. Nat Commun 10(1): 2775.
- Dietz, H., Bertz, M., Schlierf, M., Berkemeier, F., Bornschlogl, T., Junker, J. P. and Rief, M. (2006). Cysteine engineering of polyproteins for single-molecule force spectroscopy. Nat Protoc 1(1): 80-84.
- Durner, E., Ott, W., Nash, M. A. and Gaub, H. E. (2017). Post-translational sortase-mediated attachment of high-strength force spectroscopy handles. Acs Omega 2(6): 3064-3069.
- Giganti, D., Yan, K., Badilla, C. L., Fernandez, J. M. and Alegre-Cebollada, J. (2018). Disulfide isomerization reactions in titin immunoglobulin domains enable a mode of protein elasticity. Nat Commun 9(1): 185.
- He, C., Genchev, G. Z., Lu, H. and Li, H. (2012). Mechanically untying a protein slipknot: multiple pathways revealed by force spectroscopy and steered molecular dynamics simulations. J Am Chem Soc 134(25): 10428-10435.
- Hoffmann, T., Tych, K. M., Crosskey, T., Schiffrin, B., Brockwell, D. J. and Dougan, L. (2015). Rapid and robust polyprotein production facilitates single-molecule mechanical characterization of β-Barrel assembly machinery polypeptide transport associated domains. ACS Nano 9(9): 8811-8821.
- Infante, E., Stannard, A., Board, S. J., Rico-Lastres, P., Rostkova, E., Beedle, A. E. M., Lezamiz, A., Wang, Y. J., Gulaidi Breen, S., Panagaki, F., Sundar Rajan, V., Shanahan, C., Roca-Cusachs, P. and Garcia-Manyes, S. (2019). The mechanical stability of proteins regulates their translocation rate into the cell nucleus. Nat Phys 15: 973-981.
- Krieg, M., Fläschner, G., Alsteens, D., Gaub, B. M., Roos, W. H., Wuite, G. J. L., Gaub, H. E., Gerber, C., Dufrêne, Y. F. and Müller, D. J. (2019). Atomic force microscopy-based mechanobiology. Nat Rev Phys 1(1): 41-57.
- Mannix, A. J., Zhang, Z. H., Guisinger, N. P., Yakobson, B. I. and Hersam, M. C. (2018). Borophene as a prototype for synthetic 2D materials development. Nat Nanotech 13(6): 444-450.
- Ott, W., Jobst, M. A., Schoeler, C., Gaub, H. E. and Nash, M. A. (2017). Single-molecule force spectroscopy on polyproteins and receptor–ligand complexes: The current toolbox. J Stru Bio 197(1): 3-12.
- Pill, M. F., East, A. L. L., Marx, D., Beyer, M. K. and Clausen-Schaumann, H. (2019). Mechanical activation drastically accelerates amide bond hydrolysis, matching enzyme activity. Angew Chem Int Ed 58(29): 9787-9790.
- Popa, I., Berkovich, R., Alegre-Cebollada, J., Badilla, C. L., Rivas-Pardo, J. A., Taniguchi, Y., Kawakami, M. and Fernandez, J. M. (2013). Nanomechanics of HaloTag Tethers. J Am Chem Soc 135(34): 12762-12771.
- Scholl, Z. N. and Marszalek, P. E. (2018). AFM-based single-molecule force spectroscopy of proteins. Methods Mol Biol 1814: 35-47.
- Song, Y., Ma, Z., Yang, P., Zhang, X., Lyu, X., Jiang, K. and Zhang, W. (2019). Single-molecule force spectroscopy study on force-induced melting in polymer single crystals: the chain conformation matters. Macromolecules 52(3): 1327-1333.
- Walder, R., LeBlanc, M. A., Van Patten, W. J., Edwards, D. T., Greenberg, J. A., Adhikari, A., Okoniewski, S. R., Sullan, R. M. A., Rabuka, D., Sousa, M. C. and Perkins, T. T. (2017). Rapid characterization of a mechanically labile α-helical protein enabled by efficient site-specific bioconjugation. J Am Chem Soc 139(29): 9867-9875.
- Yang, R., Wong, Y. H., Nguyen, G. K. T., Tam, J. P., Lescar, J. and Wu, B. (2017). Engineering a catalytically efficient recombinant protein ligase. J Am Chem Soc 139(15): 5351-5358.
- Yuan, G., Liu, H., Ma, Q., Li, X., Nie, J., Zuo, J. and Zheng, P. (2019). Single-molecule force spectroscopy reveals that iron-ligand bonds modulate proteins in different modes. J Phys Chem Lett 10(18): 5428-5433.
- Zhang, S., Qian, H.-j., Liu, Z., Ju, H., Lu, Z.-y., Zhang, H., Chi, L. and Cui, S. (2019). Towards unveiling the exact molecular structure of amorphous red phosphorus by single-molecule studies. Angew Chem Int Ed 58(6): 1659-1663.
Article Information
Copyright
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Deng, Y., Shi, S., Zheng, B., Wu, T. and zheng, P. (2020). Enzymatic Construction of Protein Polymer/Polyprotein Using OaAEP1 and TEV Protease. Bio-protocol 10(8): e3596. DOI: 10.21769/BioProtoc.3596.
Category
Biochemistry > Protein > Synthesis
Molecular Biology > Protein > Detection
Biochemistry > Protein > Fluorescence
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









