- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Circadian Gene Profiling in Laser Capture Microdissected Mouse Club Cells
Published: Vol 10, Iss 8, Apr 20, 2020 DOI: 10.21769/BioProtoc.3590 Views: 4698
Reviewed by: Narayan SubramanianWoojong LeeAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Analyzing RNA Localization Using the RNA Proximity Labeling Method OINC-seq
Megan C. Pockalny [...] J. Matthew Taliaferro
Aug 5, 2025 2395 Views

In Vivo Retroviral Transduction of Cardiac Myofibroblasts Using Intramyocardial Injection Immediately Post-myocardial Infarction
Satsuki Ono [...] Michio Nakaya
Nov 20, 2025 1580 Views

A Highly Efficient siRNA Transfection Method in Primary Cultured Cortical Neurons
Xiaorong Wang [...] Zhaolong Zhang
Jan 20, 2026 345 Views
Abstract
Cell heterogeneity is high in tissues like lung. Research conducted on pure population of cells usually offers more insights than bulk tissues, such as circadian clock work. In this protocol, we provide a detailed work flow on how to do circadian clock study by RNA seq in laser capture micro-dissected mouse lung club cells. The method uses frozen tissues and is highly reproduciable.
Keywords: Club cellBackground
Circadian clock is found all over the body and in the lung it is shown to strongly oscillate in Club cells (Gibbs et al., 2009 and 2014). Whole lung circadian gene profiling is carried before in mice and rat (Sukumaran et al., 2011; Zhang et al., 2014). However, at least 15% of those circadian genes are coming from leukocytes (Haspel et al., 2014). It is useful but difficult to get cell type specific circadian gene profiling, given the high heterogeneity of cell population in tissues. Tissue digestion followed by flow cytometry sorting is usually to solve this problem but it is not clearly whether gene expression will change during this process. An alternative way is to use laser capture microdissection to take target cells from fixed tissues, Note that this could lead to comprised cell population purity. Specific cell population can be identified by antibody staining or anatomic information. In terms of Club cells in lung, they are known to be enriched in distal bronchiolar epithelium. A protocol showing how to do microarray using RNA from Club cells by laser capture (PixCell II Laser Capture Microdissection system) was published more than 10 years before and there are some advances in the technique (Betsuyaku and Senior, 2004; Cummings et al., 2011). Here I present a detailed protocol on circadian gene profiling in Club cell RNA by laser capture microdissection in Leica LMD6500 system. It covers from mouse tissue collection to get RNA samples ready for RNA seq library construction. Generally, to get good quality RNA, all tools and working areas should keep RNase contamination to the minimum (like using RNaseZAP solution to clean and keep sections dry in microdissection). For all the downstream steps, please refer to the published protocol by Jiajia Li, etc. (Li et al., 2015).
Materials and Reagents
- Smiths MedicalTM PortexTM Fine-Bore Polyethylene Tubing (Ref: 800/100/200, 0.58 mm ID and 0.96 mm OD)
- 23 G needles and 1 ml syringe
- PEN (polyethylene naphthalate) membrane slide (Leica, 11505158 ; Applied Biosystem, catalog number: LCM0522 ) and slide mail box
- Thin wall 0.5 ml Eppendorf tubes (Invitrogen, catalog number: Q32856 )
- Mice (C57BL or others)
- Phenobarbitone solution (for example, Pentoject, Animalcare limited, UK)
- 70% ethanol
- Dry ice or liquid nitrogen
- PBS tablet (Sigma, catalog number: P4417-100TAB )
- Sucrose
- RNaseZAP solution (Thermo Fisher, catalog number: AM9780 )
- Ethanol solutions (100%, 95%, 75%, 50%)
- Lysis buffer (supplied in kit) (Applied Biosystems, catalog number: KIT0204 )
- Picopure RNA isolation kit (Applied Biosystems, catalog number: KIT0204 )
- Qubit® RNA HS (High Sensitivity) assay kit (Invitrogen, catalog number: Q32852 )
- Agilent RNA 6000 Pico Kit (Agilent, catalog number: 5067-1511 )
- OCT (Optimal cutting temperature) compound solution (any brand, for example, Fisher Scientific, catalog number: 12678646 ) and PBS (see Recipes)
Equipment
- Qubit machine
- Light dark chamber
- Cryostat
- Forceps and scissors, fine string
- Leica Laser Microdissection Scope LMD 6500
- Tape station or Agilent 2100 Bioanalyzer
- -80 °C freezers
Procedure
- Lung tissue collection
- Entrained mice [7 day L/D (12 h light/12 h dark) followed by 1 day D/D (12 h dark/12 h dark)] are culled in D/D by either i.p. injection of phenobarbitone (200 μl/mouse) or neck dislocation. One mouse per time point is used and samples are collected every 4 h for 48 h. Once they are dead, take the bodies to the procedure area.
Notes:- In previous lung circadian gene profiling studies, the condition varies from L/D, D/D and L/L (12 h light/12 h light).
- The former method is preferred as sometimes tracheas can be damaged by neck dislocation.
- Though samples pooled from several mice (like n = 3) is used often in literature for this kind of study, the data from our study suggest that this may not be necessary. Sometimes, when mouse stock is limited, mice can be culled at different experiments (for example, only CT0-32 samples are collected in one experiment and the remaining is collected from another experiment). We don’t find big effect on the result; even tissues are collected almost 6 months apart.
- In previous lung circadian gene profiling studies, the condition varies from L/D, D/D and L/L (12 h light/12 h light).
- Spray mice with 70% ethanol and make a vertical cut in skin from abdomen to chin. Open the pleural cavity by piercing the pleural membrane and remove ribs above the lung. Lift and cut off thymus to expose trachea. The connective tissues surrounding trachea can then be peeled apart by blunt forceps.
- Use spring scissors to make a nick cut in the upper part of the trachea and insert the BAL lavage tube fitted with a 23 G needle to the trachea. Strings can be used to make a tie to hold the insert.
Note: It is not absolute necessary to do this as forceps can be used instead. But it helps in later steps during lung removal. - Fill about 0.8 ml OCT solutions [50% (v/v) OCT in PBS solution (made with DEPC H2O) with 5% sucrose] in a 1 ml syringe. And slowly inject the solution to inflate the lung through trachea.
- Remove the lung with the trachea closed with forceps or a string tie. Trim off the heart and thymus before freezing lungs in either dry ice or liquid nitrogen. For storage, the lungs covered with foil paper are put in -80 °C freezers.
Note: Samples frozen in liquid nitrogen are difficult to cut as the texture becomes very crisp. RNA quality from dry ice and liquid nitrogen frozen samples is comparable, without clear change.
- Entrained mice [7 day L/D (12 h light/12 h dark) followed by 1 day D/D (12 h dark/12 h dark)] are culled in D/D by either i.p. injection of phenobarbitone (200 μl/mouse) or neck dislocation. One mouse per time point is used and samples are collected every 4 h for 48 h. Once they are dead, take the bodies to the procedure area.
- Lung sectioning
- Take out the lungs and place them in the cryostat for 30 min before cutting.
- Mount the lung in vertical position and coronal sections are cut in 10 µm thickness, 3-4 sections per PEN membrane slide.
Note: Either RNase free or normal PEN slide can be used. For the latter, PEN membrane can be cleaned by UV light exposure in cell culture hood for 30 min or with RNaseZap. - It is all right to directly place the tissue sections into a slide from room temperature but once a section is on the slide, leave the slide in the cryostat. Examples of dissection areas are shown in the picture.
- Usually, 10 slides/mouse are cut for RNA sequence study (150-450 ng total RNA yield after microdissection.
Note: The amount depends on chosen method to construct library for sequencing. For Illumina Trueseq kit, 100 ng total RNA is the minimum amount requirement. When an amplification step can be used, much less material is required, with total RNA even in picrogram range. - Yield from 2-3 slides is enough for getting RNA in qPCR study.
- Place the slides in a closed slide mail box and store the slides in -80 °C freezers. It is recommended to finish the slides microdissection within 2 weeks.
- Laser capture microdissection
- Specimen slides were taken out of dry ice and quickly place into alcoholic solutions as below:
100% ethanol for 1 min, 75% ethanol 3 dips, 50% ethanol 3 dips, 95% ethanol 30 s, 100% ethanol 30 s, 100% ethanol 2-5 min. For ethanol solutions, avoid leaving open too long to prevent water coming in from air and change new solutions after prolonged use. Chemicals can be used to further dry the ethanol. It is essential to keep last ethanol solution 100%, as RNase can not work without water.
Notes:- Cresyl violet solution in ethanol can be used to stain sections but not necessary here. The bronchiolar structure is easy to recognise under light microscope without any staining.
- The aim of this process is to dissolve OCT in water and make the sections dry in the end to prevent RNase activity.
- Cresyl violet solution in ethanol can be used to stain sections but not necessary here. The bronchiolar structure is easy to recognise under light microscope without any staining.
- Leica LMD 6500 laser capture microdissection machine is used. After slides being dried down, they are placed in the slide holder. For tissue collection, 0.5 ml thin wall PCR tubes are used in the tissue collector, with 30 µl lysis buffer added (add DTT before use according to the kit protocol, Picopure RNA isolation kit, Life Technologies).
- Inspect the specimen to look for the bronchiolar airways under the microscope with 6.7x or 10x magnifications. Bronchiolar epithelium was cut off from junction between terminal bronchiole and alveolar ducts, proximally ~200 μm along the bronchiole. The laser setting should be tested beforehand. The microdissection view and laser setting used in our lab is shown in the picture below (Figure 1).
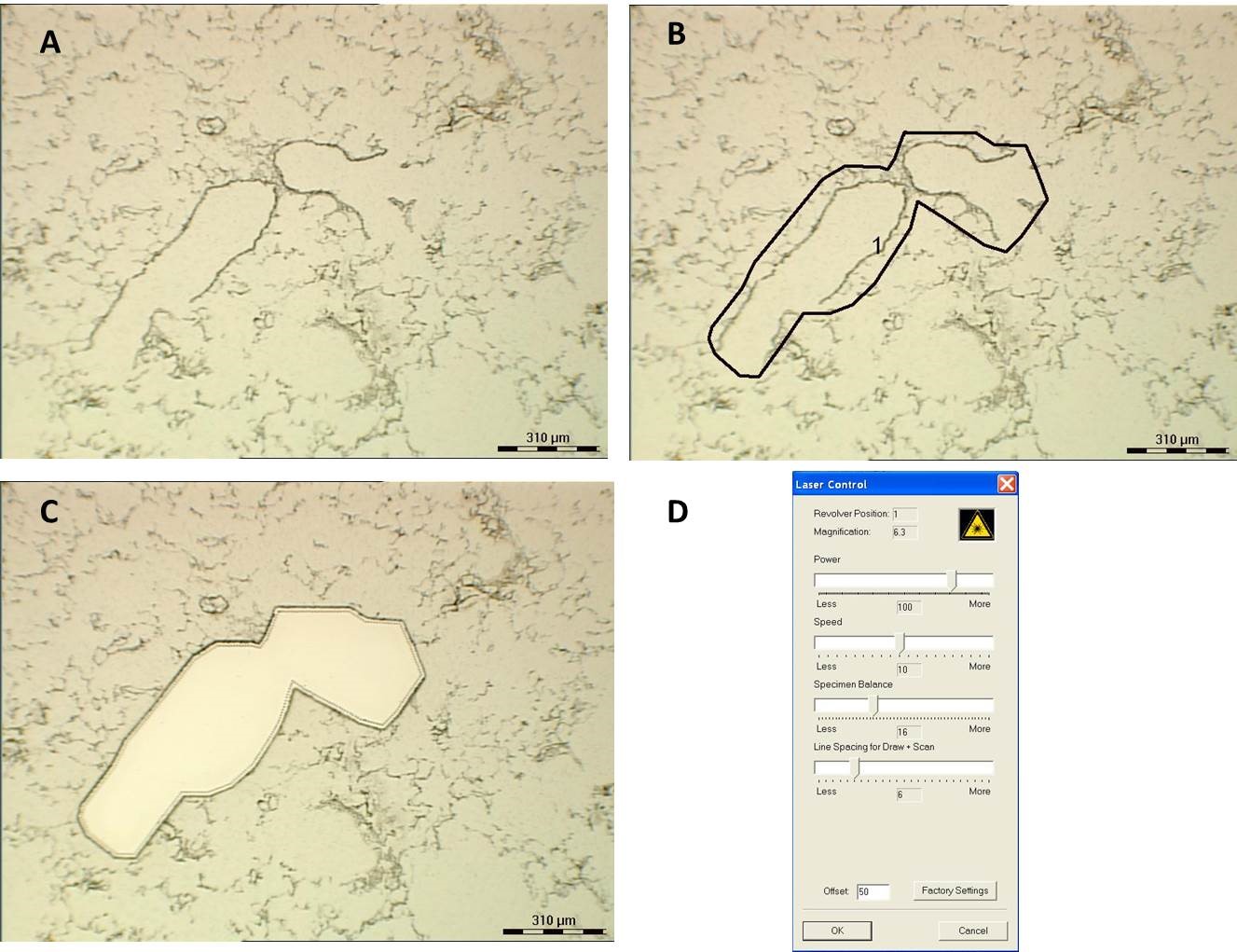
Figure 1. Illustration of laser capture of distal bronchiolar epithelial cells. The view is under 6.7x magnifications with 2 bronchiolar airways shown in the middle in A. B. shows how the cutting line is drawn and C. shows what the tissue ie like after cutting. Laser setting is shown in D. - Check the collection solution about every 1 h and add another 30 µl of lysis buffer or DEPC H2O before the solution on the cap running dry. It is recommended to use 2 or 3 collection tubes for 1 sample to avoid the solution becoming too thick after long microdissection process. These tubes for the same sample can be pooled when it comes to RNA purification.
Note: For Picopure RNA extraction kit, it is recommended to incubate samples at 42 °C for 30 min first to fully lysis the tissue. This step can be done either after immediate dissection or just before RNA extraction. - It usually takes 5-8 h to manually dissect 10 slides. In the end, put the tube onto dry ice and the samples can be stored for long term in -80 °C.
Notes:- There is an option of automatic cutting after selecting multiple target areas. However, it is not uncommon to find target sections ‘hanging’ on the slide instead of falling off after closed cutting. In this case, a manual point shooting method is used to cut off target areas.
- Each slide takes 30-45 min to microdissect. Both coarse and fine stage movement can be used during microdissection. It is recommended to inspect the sections again with eyes under microscope after dissection of each slide to avoid missing target areas.
- There is an option of automatic cutting after selecting multiple target areas. However, it is not uncommon to find target sections ‘hanging’ on the slide instead of falling off after closed cutting. In this case, a manual point shooting method is used to cut off target areas.
- Specimen slides were taken out of dry ice and quickly place into alcoholic solutions as below:
- RNA extraction and study
Picopure RNA extraction kit from Life Technologies to extract the RNA is used according to enclosed protocol. Among the extraction process, DNA on column digestion is performed as described in the protocol. Final RNA is eluted in 20 μl volume and low RNA binding Eppendorf tube is recommend to hold the samples.
Note: RNeasy micro kit from Qigen and EZNA microelute total RNA kit from VWR can also be used. It is also possible to do the extraction glycogen precipitation without any kit but it could take much longer time to do.
For quantification, it is recommended to take 1 μl of RNA and dilute 1:5 in H2O. From this, use 1 μl to quantify RNA amount in Qubit® RNA HS (High Sensitivity) assay. The typical concentration is above 10 ng/μl. The remaining can be saved for qPCR validation of final RNA seq data. For quality assessment of RNA intactness, use Agilent RNA 6000 Pico Kit or Tape station. The RIN number is usually around 6-8. If the value is too low, new mouse lung samples should be collected to start the process from beginning again.
Notes:- When the concentration is measured in a normal NanoDrop machine, the concentration can be very inaccurate. Successful qPCR application is done with laser capture microddisected materials from just 2 slides when the concentration is in lower than 0 ng/μl range by NanoDrop.
- If the amount is too low, more slides can be used to microdissect and use this RNA solution to elute or pool the RNA together in the end. Also check Step A3 about an amplification step. But it is recommended to avoid this situation by collecting as much materials as possible in first place.
- Lung is known to have high levels of RNase. In other tissues, like brain, RIN number can be as high as 8-9.
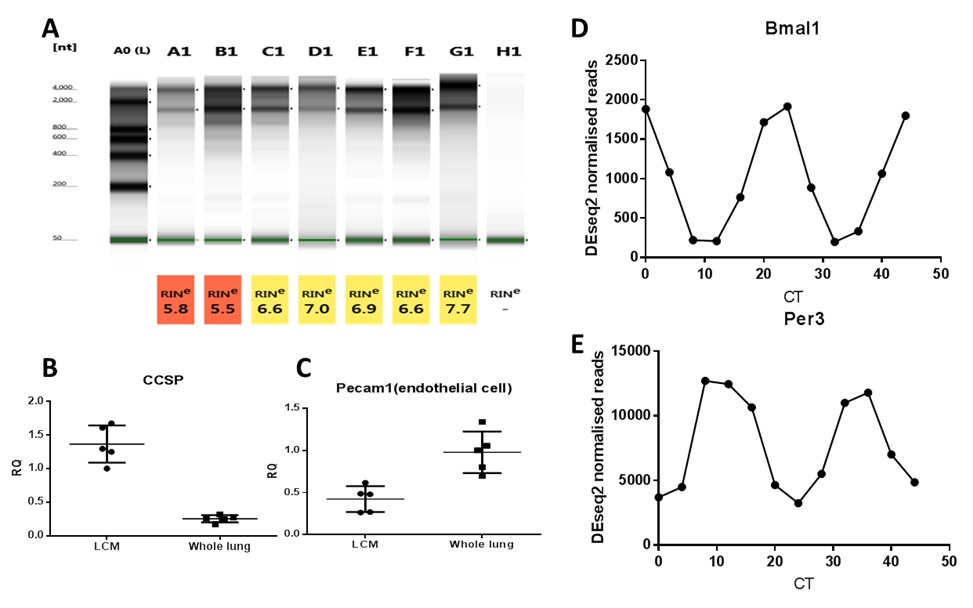
Figure 2. Typical RNA quality, qPCR and RNA seq result. A. Tape station showing RNA quality. B. and C. Enrichment of Club cell marker and little endothelial cell contamination shown by qPCR. D. and E. Bmal1 and Per3 gene expression profile from RNA seq results on laser capture microdisssected Club cells. - When the concentration is measured in a normal NanoDrop machine, the concentration can be very inaccurate. Successful qPCR application is done with laser capture microddisected materials from just 2 slides when the concentration is in lower than 0 ng/μl range by NanoDrop.
Data analysis
Study results using this method is published on FASEB journal (Zhang et al., 2019).
Recipes
- OCT (Optimal cutting temperature) compound solution and PBS
- Solutions (50% (v/v) OCT) in 1x PBS solution (made with PBS tablet in DEPC H2O) with 5% sucrose
- Dissolve sucrose first before adding OCT and this will make sucrose easier to dissolve
- Solutions (50% (v/v) OCT) in 1x PBS solution (made with PBS tablet in DEPC H2O) with 5% sucrose
Acknowledgments
The authors thank BBSRC, UK for funding of this work and the method is based on Betsuyaku and Senior (2004) and Cummings et al. (2011).
Competing interests
The authors don’t have any competing interests.
Ethics
All experiments on animals were carried out in accordance with the UK Home Office Animals (Scientific Procedures) Act of 1986 and European Directive 2010/63/EU, following approval by the University of Manchester Animal Welfare and Ethical Review Body.
References
- Betsuyaku, T. and Senior, R. M. (2004). Laser capture microdissection and mRNA characterization of mouse airway epithelium: methodological considerations. Micron 35(4): 229-234.
- Cummings, M., McGinley, C. V., Wilkinson, N., Field, S. L., Duffy, S. R. and Orsi, N. M. (2011). A robust RNA integrity-preserving staining protocol for laser capture microdissection of endometrial cancer tissue. Anal Biochem 416(1): 123-125.
- Gibbs, J. E., Beesley, S., Plumb, J., Singh, D., Farrow, S., Ray, D. W. and Loudon, A. S. (2009). Circadian timing in the lung; a specific role for bronchiolar epithelial cells. Endocrinology 150(1): 268-276.
- Gibbs, J., Ince, L., Matthews, L., Mei, J., Bell, T., Yang, N., Saer, B., Begley, N., Poolman, T., Pariollaud, M., Farrow, S., DeMayo, F., Hussell, T., Worthen, G. S., Ray, D. and Loudon, A. (2014). An epithelial circadian clock controls pulmonary inflammation and glucocorticoid action. Nat Med 20(8): 919-926.
- Haspel, J. A., Chettimada, S., Shaik, R. S., Chu, J. H., Raby, B. A., Cernadas, M., Carey, V., Process, V., Hunninghake, G. M., Ifedigbo, E., Lederer, J. A., Englert, J., Pelton, A., Coronata, A., Fredenburgh, L. E. and Choi, A. M. (2014). Circadian rhythm reprogramming during lung inflammation. Nat Commun 5: 4753.
- Li, J., Grant, G. R., Hogenesch, J. B. and Hughes, M. E. (2015). Considerations for RNA-seq analysis of circadian rhythms. Methods Enzymol 551: 349-367.
- Sukumaran, S., Jusko, W. J., Dubois, D. C. and Almon, R. R. (2011). Light-dark oscillations in the lung transcriptome: implications for lung homeostasis, repair, metabolism, disease, and drug action. J Appl Physiol (1985) 110(6): 1732-1747.
- Zhang, R., Lahens, N. F., Ballance, H. I., Hughes, M. E. and Hogenesch, J. B. (2014). A circadian gene expression atlas in mammals: implications for biology and medicine. Proc Natl Acad Sci U S A 111(45): 16219-16224.
Article Information
Copyright
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Zhang, Z. and Loudon, A. S. I. (2020). Circadian Gene Profiling in Laser Capture Microdissected Mouse Club Cells. Bio-protocol 10(8): e3590. DOI: 10.21769/BioProtoc.3590.
Category
Systems Biology > Transcriptomics > RNA-seq
Cell Biology > Cell-based analysis > Gene expression
Molecular Biology > RNA > RNA sequencing
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link