- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
An in vitro Assay to Screen for Substrates of PKA Using Phospho-motif-specific Antibodies
Published: Vol 10, Iss 8, Apr 20, 2020 DOI: 10.21769/BioProtoc.3587 Views: 4222
Reviewed by: Gal HaimovichNathalie EisenhardtAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Endo-1,4-β-D-xylanase Assay Using Azo-Xylan and Variants Thereof
Luca Bombardi [...] Salvatore Fusco
Apr 20, 2025 1916 Views

Activation of X-Succinate Synthases for Fumarate Hydroalkylation Using an In Vitro Activation Method
Anshika Vats [...] Mary C. Andorfer
Jun 20, 2025 2510 Views

An Optimized Enzyme-Coupled Spectrophotometric Method for Measuring Pyruvate Kinase Kinetics
Saurabh Upadhyay
Aug 20, 2025 2441 Views
Abstract
Kinases function as regulators of many cellular processes such as cell migration. These enzymes typically phosphorylate target motif sequences. Mass spec or phospho-specific antibody detection can be used to determine whether a kinase can phosphorylate proteins of interest, however, mass spec can be expensive and phospho-antibodies for the protein of interest may not exist. In this protocol, we will describe an in vitro kinase assay to provide a preliminary readout on whether a protein of interest may be phosphorylated by PKA. Our protein of interest is purified after expression in bacteria and treated with recombinant PKA from bovine heart. Protein is then extracted and a western blot is performed using a phospho-specific antibody for PKA’s target motif. This will allow us to quickly determine if it is possible for PKA to phosphorylate our protein of interest.
Keywords: Protein Kinase A (PKA)Background
Hormones and other factors that cause activation of adenylate cyclase through G-linked G-protein coupled receptors (GPCR) and subsequently promote generation of second messenger cAMP can affect cellular processes like cell migration. Elevation of cAMP level leads to activation of PKA, a serine-threonine kinase that plays an important role in the regulation of actin cytoskeletal dynamics in migrating cells. PKA influences different facets of actin cytoskeleton-regulatory processes including modulation activities of a) Rho-family GTPases (Rho, Rac and Cdc42), b) actin-binding proteins (e.g., VASP [vasodilator stimulated phosphoprotein]), c) kinases which indirectly control the function of actin-binding proteins (e.g., p21-activated kinase) and d) myosin (Howe, 2004). PKA and other kinases, however, can also modulate other proteins by phosphorylation that are currently unknown.
There are multiple ways to determine kinase activity on a protein of interest such as mass spec for phosphopeptides, however, such assays can be costly. In this protocol, we will describe the use of an in vitro kinase assay to examine PKA kinase activity on a GST-tagged protein of interest using a phospho-specific antibody for PKA’s target motif. Using a PKA motif antibody is advantageous if known phosphorylation sites are unknown on a protein or if no phosphospecific protein antibody exists. While this assay is quick and simple to perform, it depends on the kinase targeting a motif pre-determined by available antibodies and thus limited in potential. A different kinase can be easily tested if an antibody to detect that kinase’s phospho motif exists. Despite this, successful readout from this assay can provide motivation to pursue more comprehensive exploration into kinase phosphorylation of a protein of interest. Although this protocol describes usage of recombinant protein purified from bacteria, protein can be obtained from any source (i.e., insect or human cells) but total yield of protein will vary dependent on the source and may need to be optimized. Figure 1 below illustrate the general workflow for this protocol.

Figure 1. General protocol workflow for kinase analysis
Materials and Reagents
- Eppendorf tubes
- Bacteria culture tube (Southern Labware, catalog number: 110178 )
- BL21 competent E. coli (New England BioLabs, catalog number: C2530H )
- LB Broth (Thermo Fisher, catalog number: 10855001 )
- LB Agar Ampicillin-100 (Sigma-Aldrich, catalog number: L5667 )
- IPTG (Sigma-Aldrich, catalog number: I6758 )
- Ampicillin 100 mg/ml (Sigma-Aldrich, catalog number: A5354 )
- Glutathione Agarose (Thermo Fisher, Pierce, catalog number: 16100 )
- Protease inhibitor cocktail for bacteria (Sigma-Aldrich, catalog number: P8465 )
- pGEX-4T1 GST-fusion vector (Sigma-Aldrich, catalog number: GE28-9545-49 )
- PBS (Lonza, BioWhittaker, catalog number: BW17516F )
- IP Lysis Buffer (Thermo Fisher, Pierce, catalog number: 87787 )
- PKA (Sigma-Aldrich, catalog number: P2645-400UN )
- Laemmli SDS sample buffer, reducing 6x (Alfa Aesar, catalog number: J61337-AC )
- BES (Thermo Fisher, BioReagents, catalog number: BP501500 )
- EGTA (Tocris Bioscience, catalog number: 28-071-G )
- MgCl2 (Thermo Fisher, catalog number: AB0359 )
- ATP (Sigma-Aldrich, catalog number: A7699 )
- Phosphocreatine (Sigma-Aldrich, catalog number: P7936 )
- DTT (Sigma-Aldrich, catalog number: D9779 )
- Prestained protein ladder (Thermo Fisher, catalog number: 26616 )
- 15% Criterion Tris-HCl protein gel (Bio-Rad, catalog number: 3450019 )
- 10x Tris/Tricine/SDS running buffer (Bio-Rad, catalog number: 1610744 )
- 10x Tris/Glycine transfer buffer (Bio-Rad, catalog number: 1610734 )
- Methanol (Thermo Fisher, Fisher Chemical, catalog number: A412-500 )
- Nitrocellulose membrane (Bio-Rad, catalog number: 1620115 )
- TBS-T (Bio-Rad, catalog number: BUF028 )
- BSA (Thermo Fisher, BioReagents, catalog number: BP1600100 )
- Phospho-PKA subtract RRXS antibody (Cell Signaling Technology, catalog number: 9624 )
- Peroxidase Goat Anti-Rabbit IgG (Jackson ImmunoResarch, catalog number: 111-035-144 )
- Clarity ECL (Bio-Rad, catalog number: 1705060 )
- Kinase buffer (see Recipes)
- IPTG solution (see Recipes)
Equipment
- Refrigerated Centrifuge for Eppendorf tubes (Sorvall, Legend RT, SO-LEGRT)
- Eppendorf tube heater (Lab Line, Multi Blok Heater, LV40429530)
- Shaking incubator set to 37 °C (Thermo Scientific, MaxQ 5000, SHKE50007)
- Waterbath or drybath set to 37 °C (Boekel Scientific, Small Water Bath, 290100)
- Sonicator (Bransonic, Ultrasonic Baths, CPX-952-116R)
- Rotator for Eppendorf tube (Boekel Scientific, Scientific Rotating Tube Rotator, UX-51202-00)
- Microcentrifuge (Thermo Scientific, accuSpin Micro 17, 13-100-676)
- Mini Trans-Blot (Bio-Rad)
- Mini-PROTEAN Tetra cell (Bio-Rad)
- ChemiDoc XRS+ System (Bio-Rad)
Software
- ImageLab (Bio-Rad)
Procedure
- Transformation
- Thaw BL21 competent cells on ice.
- Combine 50 μl of BL21 cells with 100 ng of plasmid that can be expressed/induced in bacteria (i.e., contains laclq gene sequence which prevents expression until induced by IPTG), in our case, GST vector (pGEX-4T1) with gene of interest (in our case, Profilin-1) and incubate on ice for 30 min.
- Heat shock mixture at 42 °C for 45 s using water bath or heat block.
- Place mixture back on ice for 2 min.
- Incubate bacteria mixture in 200 μl of LB broth in shaking incubator for 1 h at 300 rpm and 37 °C (make sure mixture is not in an airtight tube).
- Plate all the mixture onto LB agar plate containing appropriate antibiotics, in our case, ampicillin (final concentration 100 μg/ml) and spread until plate is dry.
- Incubate at 37 °C overnight (ensure plate is inverted).
- Protein induction and extraction
- Pick a single colony from bacteria plate and grow in 3 ml of LB broth and appropriate antibiotic overnight (ampicillin final concentration 100 μg/ml) in bacteria culture tube in shaking incubator at 37 °C (ensure tube is not airtight).
- Take 1 ml of this solution and grow in 44 ml of LB broth and appropriate antibiotic (ampicillin final concentration 100 μg/ml) at 37 °C in rotating incubator at 300 rpm for 3 h.
- Add 1:1,000 100 mM IPTG to induce expression to mixture and incubate at 300 rpm for another 3 h.
- Transfer bacteria solution into a 50 ml centrifuge tube and centrifuge bacteria culture for 20 min at 3,500 x g and 4 °C to harvest bacteria.
- Discard supernatant.
- Resuspend bacteria pellet in 850 μl of IP lysis buffer with 50 μl/ml of bacterial protease inhibitor and transfer into a microcentrifuge Eppendorf tube. Bacterial protease inhibitor should be prepared and used fresh each time.
- Sonicate for 10 s (no heat), rest on ice for 2 min, and repeat 2 more times. This step is used to shear DNA and break bacteria.
- Centrifuge at 17,000 x g for 30 min at 4 °C.
- Collect supernatant.
Note: Product can be run on a gel and Coomassie stained to confirm protein expression.
- Protein purification and preparation for kinase treatment
- Aliquot ~400 μl of Glutathione Agarose (GST bead slurry) to Eppendorf tubes.
- Wash two times with 500 μl cold PBS and centrifuge beads down between washes at 8,000 x g for 1 min.
- Wash one time with 500 μl IP lysis buffer and centrifuge beads down at 8,000 x g for 1 min.
- Add IP lysis buffer to make 50/50 slurry of beads and IP lysis buffer.
- Transfer 160 μl of this slurry into a new Eppendorf tube and add 540 μl of IP lysis buffer to it and 50 μl/ml bacterial protease inhibitor. Bacterial protease inhibitor should be prepared and used fresh each time.
- Add 300 μl of bacteria lysate and rotate using rotating end-over-end rotator at 30 rpm for 2 h at 4 °C.
- Centrifuge beads down at 8,000 x g for 1 min and wash three times with 500 μl IP lysis buffer and centrifuge beads down between washes at 8,000 x g for 1 min.
- Wash two times with 500 μl kinase buffer and centrifuge beads down between washes at 8,000 x g for 1 min (leave at 50/50 slurry).
- Kinase treatment
- Reconstitute PKA by adding 200 μl of kinase buffer (400UN of PKA comes as a dry pellet) and let sit for 20 min at room temp, this is your PKA solution.
- Aliquot 2 Eppendorf tubes with 40 μl of GST bead solution containing protein of interest from previous section. One tube will be used as a negative control.
- Add 5 μl of PKA solution to one tube (aliquot the rest of the PKA solution into 5 μl and store at -80 °C for future use) and 5 μl of kinase solution (i.e., kinase solution with no PKA) to the other. Note that PKA is susceptible to freeze-thaw and loses some activity after the first freeze. The recommended aliquot amount is enough for one treatment group each time and thus no need to freeze-thaw. PKA should not be used after being frozen for > 1 year in -80 °C.
- Incubate at 32 °C for 1 h using water bath or heat block and mix using vortex every 15 min to ensure beads are not settled to bottom of tube.
- Wash two times with 500 μl IP lysis buffer buffer and centrifuge beads down between washes at 8,000 x g for 1 min and leave at 50/50 slurry.
- Aliquot 30 μl of solution to a new tube and add 60 μl of 1.5x Laemilli buffer.
- Boil for 5 min and centrifuge at 17,000 x g for 5 min.
- Transfer supernatant to a fresh microcentrifuge Eppendorf tube.
- Western blot
- Load 5 μl of protein ladder into 15% gel and load 30 μl of protein sample (supernatant from last step in previous section) into adjacent wells.
- Fill PROTEAN with 1x running buffer and run at 200 V for ~45 min (might be more or less time, run until blue dye from sample buffer runs off the bottom of the gel). These settings were used for our protein-of-interest, settings may be different for other proteins.
- Transfer protein from gel onto either PVDF or nitrocellulose membrane (we used nitrocellulose) by using Trans-Blot filled with 1x transfer buffer (80% transfer buffer, 20% methanol) at 100 V for ~1.5 h (might be more or less depending on size of protein of interest). Make sure gel and membrane are placed in the correct orientation, i.e., the gel should be closer to the negative charge source (typically the black colored part of the transfer cassette should be closest to the gel). These settings were used for our protein-of-interest, settings may be different for other proteins.
- Cut membrane around protein of interest (1-2 ladder bands above and below protein weight) and block with 5% BSA in TBS-T for 30 min at room temperature with rocking motion. We used an empty microscope coverslip slide box to incubate our membranes (total blocking buffer volume 1.5 ml). In our case, GST-Pfn1 has a molecular weight of ~41 kd, so membrane was cut between 30 and 70 kd. Depending on the mass of your protein-of-interest, a different section may need to be cut based off the protein ladder.
- Incubate overnight 5% BSA in TBS-T with 1:1,000 pRRXS substrate antibody at 4 °C with rocking motion at 10 rpm. Using the same vessel as described previously, we use 1.5 ml of blocking solution and 1.5 μl of antibody.
- Wash with TBS-T one time for 5 min.
- Incubate with 5% BSA in TBS-T for 1 h at room temp with 1:1,000 anti-rabbit peroxidase antibody with rocking motion at 10 rpm. Using the same vessel as described previously, we use 1.5 ml of blocking solution and 1.5 μl of antibody.
- Wash 3-4 times with TBS-T for 5 min each wash.
- Add Clarity ECL solution to membrane and incubate for 1 min at room temperature.
- Image membrane (example results shown in Figure 2, PKA treated GST-Pfn1 is detected by pRRXS antibody, using a BioRad ChemiDoc).
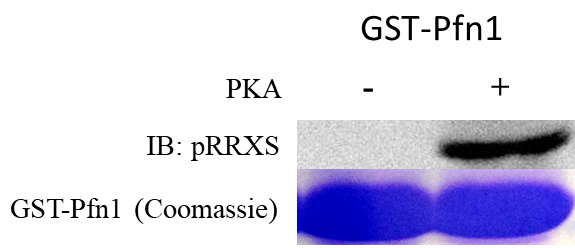
Figure 2. pRRXS antibody detects PKA phosphorylated GST-Pfn1. GST-Pfn1 is phosphorylated by PKA and detected by pRRXS antibody whereas kinase buffer solution treated GST-Pfn1 (negative control) is not detected. A Coomassie from this sample is shown as well to illustrate the amount of pulldown to obtain the phospho-band.
Data analysis
- Compare lane from protein sample treated with PKA and sample not treated with PKA.
- If band intensity is higher in PKA treated sample, the sample has increased phosphorylation as per pRRXS phosphospecific PKA antibody (in our case, in Figure 2, we saw no band intensity to full band intensity after treatment with PKA).
Recipes
- Kinase buffer
Final concentration of all components is listed below:
20 mM BES pH 7 (starts as powder, dilute in water and pH correct until appropriate concentration). This is used as the base of the Kinase buffer
20 mM EGTA (starts as powder, add into BES solution)
6 mM MgCl2 (starts as powder, add into BES solution)
5 mM ATP (starts as powder, add into BES solution)
10 mM phosphocreatine (starts as powder, add into BES solution)
1 mM DTT (starts as powder, add into BES solution) - IPTG solution
Make 100 mM IPTG solution using water and IPTG powder and store at -20 °C
Acknowledgments
This work was supported by a grant from the National Institute of Health (2R01CA108607) to PR. David Gau was supported by a National Science Foundation pre-doctoral fellowship (2012139050) and an NIH Cardiovascular Bioengineering pre-doctoral training grant (2T32HL076124 to DG).
Competing interests
The authors have no competing interests to report.
References
- Howe, A. K. (2004). Regulation of actin-based cell migration by cAMP/PKA. Biochim Biophys Acta 1692(2-3): 159-174.
Article Information
Copyright
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Gau, D. M. and Roy, P. (2020). An in vitro Assay to Screen for Substrates of PKA Using Phospho-motif-specific Antibodies . Bio-protocol 10(8): e3587. DOI: 10.21769/BioProtoc.3587.
- Gau, D., Veon, W., Shroff, S. G. and Roy, P. (2019). The VASP-profilin1 (Pfn1) interaction is critical for efficient cell migration and is regulated by cell-substrate adhesion in a PKA-dependent manner. J Biol Chem 294(17): 6972-6985.
Category
Molecular Biology > Protein > Phosphorylation
Cell Biology > Cell-based analysis > Enzymatic assay
Biochemistry > Protein > Activity
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link