- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Fibrin Breakdown Assay
Published: Vol 10, Iss 8, Apr 20, 2020 DOI: 10.21769/BioProtoc.3585 Views: 5422
Reviewed by: Antoine de MorreeStéphane RomeroSurabhi Sonam

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Protein Turnover Dynamics Analysis With Subcellular Spatial Resolution
Lorena Alamillo [...] Edward Lau
Aug 5, 2025 2397 Views

Immunopeptidomics Workflow for Isolation and LC-MS/MS Analysis of MHC Class I-Bound Peptides Under Hypoxic Conditions
Hala Estephan [...] Eleni Adamopoulou
Nov 20, 2025 1846 Views

Quantitative Proteomics of Nitrosylated Proteins in Melanoma Using the Biotin-Switch Technique Combined With Tandem Mass Tag Labeling
Vipin K. Yadav [...] Sanjay Premi
Dec 5, 2025 1539 Views
Abstract
Fibrinolysis is an integral part of the matrix remodeling process that contributes to tissue repair. Fibrin clots are broken down during fibrinolysis in a controlled process. Fibrin degradation products (FDPs) have also been shown to have a role in the regulation of cell growth and are implicated in various vascular diseases. This protocol was designed to quantitatively measure the extent of fibrin breakdown and how this can be adapted as a tool to further investigate the pathway involved in fibrinolysis or fibrin degradation products. Until now, we haven’t found an alternative method to analysis fibrinolysis.
Keywords: FibrinBackground
In hemostasis, fibrin plays a crucial role as both the ultimate substrate for fibrinolysis and the primary product of the coagulation cascade (Cesarman-Maus and Hajjar, 2005; Chapin and Hajjar, 2015). In addition, fibrin has been found to mediate the acute inflammatory response to implanted biomaterials and it has been suggested that fibrin plays a prominent role in leukocyte transmigration and thereby inflammation (Yakovlev and Medved, 2018). Fibrinolysis is a tightly regulated process by which a fibrin-rich thrombus is formed and degraded, to prevent blood clots from growing (Amengual and Atsumi, 2016) and to contribute to tissue repair (Houslay et al., 2019). In Fibrinolysis, a fibrin clot (the product of coagulation) is broken down. Fibrin degradation products (FDPs), fibrous proteins that act as mitogenic factors, can promote the proliferation of endothelial cells, smooth muscle cells and fibroblasts, and cholesterol deposition (Dong et al., 2017). Numerous studies have shown that FDPs are involved in various vascular diseases. For example, FDPs induce the adhesion and gathering of leucocytes, damaging blood vessel endothelium (Dong et al., 2017). Also, FDPs have been approved that have diverse effects in inflammatory processes and acute phase responses, such as activating TLR-4 and integrin and cell adhesionor migration (Schuliga, 2015). They inhibit several neutrophil functions crucial to the bactericidal role of these inflammatory cells (Gerdin et al., 1980). It is vital that a more complete understanding of the mechanisms behind the FDPs formation and related pathway is gained. In sight of this, we present an assay called the Fibrin breakdown assay to use in laboratory research. This assay is designed to measure the extent of fibrin degradation quantitatively. Until now, we have found no alternative method to analysis fibrinolysis.
Materials and Reagents
- 24-well cell culture plates (Corning®, catalog number: 3524-SLS )
- TC Dish 100 mm (Corning®, catalog number: 430167-SLS )
- TC Plate 6-well (Corning®, catalog number: 3516-SLS )
- 50 ml Falcon tube
- 12-well plate
- NIH3T3 cells: fibroblasts from Swiss mouse embryo tissue with the ‘3T3’ designation referring to the abbreviation of “3-day transfer, inoculum 3 x 105 cells”
- Mouse embryonic Fibroblast (MEF) cells: isolated from the bodies of c57 black mouse embryos
- Dulbecco’s modified Eagle’s medium (500 ml) DMEM HIGH GUCOSE no glutamine/sod.pyruvate (Sigma-Aldrich, catalog number: D5671 )
- Phosphate Buffered Saline (PBS) 1 tablet/200 ml (Sigma-Aldrich, catalog number: P4417-100TAB )
- Trypsin EDTA (1x) (Gibco, catalog number: 15400-054 )
- Thrombin (Sigma-Aldrich, catalog number: 10602400001 )
- Factor XIII (Sigma-Aldrich, catalog number: 233501-100UG-M )
- Fibrinogen (Sigma-Aldrich, catalog number: F3879-100MG )
- Na-citrate (Sigma-Aldrich, catalog number: 234265-100G )
- Sodium chloride (VWR CHEMICALS, catalog number: 27810.295P )
- Plasminogen (Sigma-Aldrich, catalog number: P7999-5UN )
- Newborn Calf Serum (NCS) (Sigma-Aldrich, catalog number: 12023C-500ML )
- Penicillin/Streptomycin (Sigma-Aldrich, catalog number: P0781-100ML )
- L-glutamine (Sigma-Aldrich, catalog number: G7513-100ML )
- Complete Dulbecco’s modified Eagle’s medium (DMEM) (see Recipes)
- Fibrin Matrices (see Recipes)
Equipment
- TC Flasks 150 cm vented (Corning®, catalog number: CLS430825-50EA )
- Analytical Balance (Sartorius, catalog number: CP124S )
- Phase contrast microscope (Zeiss)
- Cell culture incubator (Thermo Scientific Heraeus®)
- Centrifuge (MSE Mistral 1000)
Software
- GraphPad Prism 7 (GraphPad, https://www.graphpad.com/)
- Excel (Microsoft, https://www.microsoft.com/en-ca/)
- ImageJ bundled with 64-bit Java 1.8.0 (NIH, https://imagej.nih.gov/ij/)
Procedure
- Maintenance of cell lines
- Progate the cells in growth media containing Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 0.1% penicillin/streptomycin (10,000 U/ml), 2 mM glutamine and 10% newborn calf serum (NCS).
- The cells were passaged when approximately 70-90% confluence was reached.
- To passage the cells, remove the growth media and add 5 ml of sterile pre-warmed phosphate buffer saline (PBS) to gently wash the cells to get rid of the remaining growth media.
Note: Remaining media will reduce the efficiency of trypsin. - Remove the PBS and add 5 ml of trypsin-EDTA solution. Incubate the cells for 5 min at 37 °C.
- Agitate the cells vigorously and then analyze the cells under the microscope to ensure complete cell detachment.
- Add 5 ml of growth media containing NCS to inactivate the trypsin-EDTA solution.
- Collect the cells by centrifugation at 80 x g for 3 min at room temperature.
- Remove the growth media and trypsin-EDTA solution. Re-suspend the cell pellet in 5 ml of fresh growth media.
- Once fully resuspended, add 1 ml of cells to fresh growth media in sterile flasks, 100 mm dishes or six well plates.
- Incubate the cells at 37 °C in an atmosphere of 95% humidity, 5% CO2 and ambient O2 until required or confluent.
- Fibrin matrices preparation
- Add 0.1 U/ml thrombin into a 50 ml Falcon tube with a mixture of 2.5 U factor XIII, 2 mg fibrinogen, 2 mg Na-citrate, 0.8 mg NaCl, and 3 μg plasminogen per ml DMEM medium without indicator (Lansink et al., 1998).
- Add 300 μl of the mixture into each well of a 12-well plate. Let mixture clot at room temperature.
- Cell seeding and measurement
- Add 0.5 ml DMEM supplemented with 10% NCS and 1% penicillin/streptomycin.
- Weigh out the fibrin matrices with 12-well plate before adding medium and seeding cells.
- Seed the cells at 1 x 105 per well of a 12-well plate containing fibrin matrices, in order to obtain confluent monolayers cultured in DMEM (without indicator) supplemented with 10% NCS, 2 mM -glutamine and 1% penicillin/streptomycin.
- Incubate the plates for 8-12 days at the same environment as described above.
- Add test compounds together with fresh medium every 2-3 days.
- Collect the medium and replace with new media every 2 to 3 days along with new test compounds.
- Use phase contrast microscope to analyze invading cells in three-dimensional fibrin matrix, take 10x magnification images of the fibrin gels and lytic zones.
- Aspirate the fibrin degradation supernatant, weigh out the remaining gel with 12-well plate using an analytical balance.
- Decrease in gel weight corresponds to increased fibrin degradation.
Data analysis
- Use phase contrast microscopy to analyze cell invasion into the 3D fibrin matrix.
- To quantify fibrin degradation, the supernatant was aspirated, cells were removed before weighing, and the remaining gel was weighed using an analytical balance. Decrease of gel weight corresponded to increased fibrin gel degradation.
- Values are presented as mean ± S.E.M. from at least three independent experiments. Statistically significant between groups were determined using Student’s t-test. Values were considered significant if P < 0.05.
- A set of images and statistic analysis:
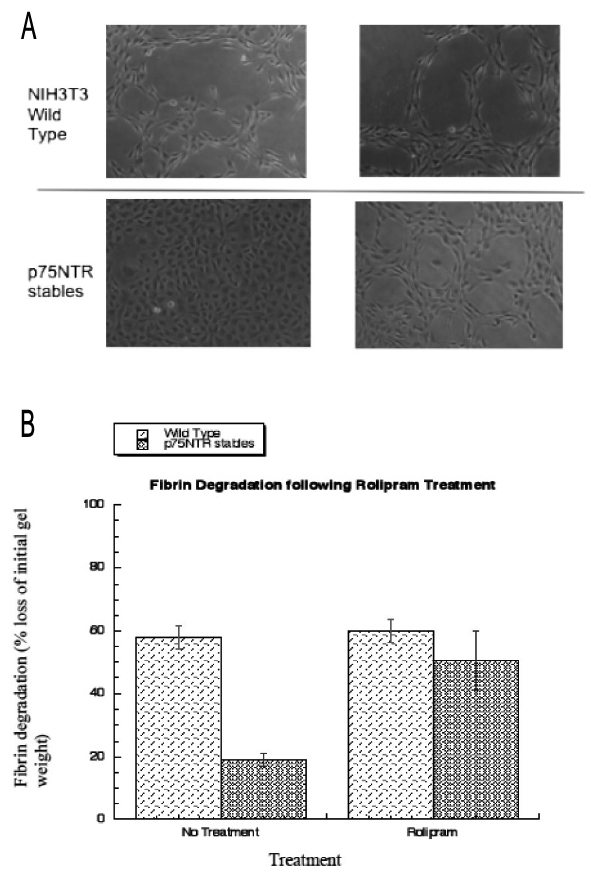
Figure 1. Expression of p75NTR regulates fibrinolysis in NIH3T3 fibroblasts and is PDE4 dependent. NIH3T3 cells and NIH3T3 p75NTR stable cells were seeded into 3-Dimensional fibrin gels and incubated at 37 °C for 9 days. Cells were either left untreated or subjected to prolonged treatment with the PDE4-specific inhibitor, rolipram (10 μM) for 8 days. A is 10x magnification images of fibrin gels and lytic zones. B is quantification of degradation of the 3D fibrin gels calculated by weighing after 9 days. All data shown are representative mean data ± standard deviation of three separate experiments.
Notes
The negative control and positive control condition should be considered, a well with fibrin matrix but no cells, or a well with cells but no fibrin matrix, and treated under the same treatment for negative control; fibrin matrices + cells + pro-fibrin degradation compound(s) that have been validated in literature, or fibrin matrix + diseased-vascular cells that are known to become fibrotic as a positive control.
Recipes
- Complete Dulbecco’s modified Eagle’s medium (DMEM)
10% newborn calf serum (NCS)
0.1% penicillin/streptomycin (10,000 U/ml)
2 mM glutamine in 500 ml DMEM
Store at 4 °C - Fibrin Matrices
0.1 U/ml thrombin
2.5 U factor XIII
2 mg fibrinogen
2 mg Na-citrate
0.8 mg NaCl
3 µg plasminogen in per ml comlplete DMEM growth media
Acknowledgments
Jiayue Ling is funded by the China Scholarship Council. We thank Kirsty F. Houslay for developing this assay. This protocol described herein were originally described in Houslay et al. (2019).
Competing interests
The authors declare that they have no conflicts of interest with the contents of this article.
References
- Amengual, O. and Atsumi, T. (2016). Chapter 56: Pathogenesis of Antiphospholipid Syndrome. Systemic Lupus Erythematosus 487-494.
- Cesarman-Maus, G. and Hajjar, K. A. (2005). Molecular mechanisms of fibrinolysis. Br J Haematol 129(3): 307-321.
- Chapin, J. C. and Hajjar, K. A. (2015). Fibrinolysis and the control of blood coagulation. Blood Rev 29(1): 17-24.
- Dong, J., Duan, X., Feng, R., Zhao, Z., Feng, X., Lu, Q., Jing, Q., Zhou, J., Bao, J. and Jing, Z. (2017). Diagnostic implication of fibrin degradation products and D-dimer in aortic dissection. Sci Rep 7: 43957.
- Gerdin, B., Saldeen, T., Roszkowski, W., Szmigielski, S., Stachurska, J. and Kopec, M. (1980). Immunosuppressive effect of vasoactive peptides derived from human fibrinogen. Thromb Res 18(3-4): 461-468.
- Houslay, K. F., Fertig, B. A., Christian, F., Tibbo, A. J., Ling, J., Findlay, J. E., Houslay, M. D. and Baillie, G. S. (2019). Phosphorylation of PDE4A5 by MAPKAPK2 attenuates fibrin degradation via p75 signalling. J Biochem 166(1): 97-106.
- Lansink, M., Koolwijk, P., van Hinsbergh, V. and Kooistra, T. (1998). Effect of steroid hormones and retinoids on the formation of capillary-like tubular structures of human microvascular endothelial cells in fibrin matrices is related to urokinase expression. Blood 92(3): 927-938.
- Schuliga, M. (2015). The inflammatory actions of coagulant and fibrinolytic proteases in disease. Mediators Inflamm. doi: 10.1155/2015/437695.
- Yakovlev, S. and Medved, L. (2018). Effect of fibrinogen, fibrin, and fibrin degradation products on transendothelial migration of leukocytes. Thromb Res 162: 93-100.
Article Information
Copyright
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Ling, J., Blair, C. M. and Baillie, G. S. (2020). Fibrin Breakdown Assay. Bio-protocol 10(8): e3585. DOI: 10.21769/BioProtoc.3585.
Category
Biochemistry > Protein > Degradation
Biochemistry > Protein > Quantification
Cell Biology > Cell-based analysis > Enzymatic assay
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









