- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Isolation, Purification and Characterization of Exosomes from Fibroblast Cultures of Skeletal Muscle
Published: Vol 10, Iss 7, Apr 5, 2020 DOI: 10.21769/BioProtoc.3576 Views: 7556
Reviewed by: Alka MehraKristina Aguilerailgen Mender

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Primary Neuronal Culture and Transient Transfection
Shun-Cheng Tseng [...] Eric Hwang
Jan 20, 2025 2789 Views

Isolation and Characterization of Cervical Cancer-Associated Mesenchymal Stem Cells From Primary Tumors Using Explant Culture
Surbhi Singla [...] Shalmoli Bhattacharyya
Jun 20, 2025 2798 Views

Generation of Intestinal Epithelial Monolayers From Single-Cell Dissociated Organoids
Neta Felsenthal and Danijela Matic Vignjevic
Oct 5, 2025 2349 Views
Abstract
Exosomes are dynamic nanovesicles secreted by virtually all cells and are present in all biological fluids. Given their highly heterogeneous content exosomes have been implicated in many physiological and pathological processes that they exert by influencing cell-cell and cell-ECM communication. In recent years an increasing number of methods have been established for the purification and characterization of exosomes. These include ultracentrifugation, ultrafiltration, size exclusion chromatography, immune capture and precipitation using a proprietary polymer. Here, we provide a protocol based on differential ultracentrifugation and sucrose density gradients tailored for the isolation of crude and ultra-pure exosomes from primary fibroblast cultures derived from adult mouse skeletal muscle. This protocol can be adapted and modified for the isolation and characterization of exosomes from a variety of tissues and bodily fluids.
Keywords: FibroblastsBackground
Exosomes are single-membrane, heterogeneous nanovesicles ranging in diameter from 30 to 150 nm, secreted by all cells and present in virtually all body fluids. The spectrum of soluble and membrane macromolecules, mRNAs, microRNAs present in exosomes depends on the metabolic status as well as the developmental origin of the cell(s) secreting these nanovesicles. Owing to their cargo composition, exosomes can initiate signaling pathways in the receiving cells and have been involved in development, immunity and in the maintenance of normal tissue physiology. In disease conditions, like neurodegeneration, fibrosis and cancer, they have been shown to trigger and propagate pathological stimuli (Rackov et al., 2018; Gurunathan et al., 2019; van de Vlekkert et al., 2019). Here we describe a protocol for the purification of exosomes from fibroblast cultures established from adult mouse gastrocnemius (GA) muscle (van de Vlekkert et al., 2019). These purified exosomal preparations can be subjected to mass spectrometry. The advantage of this method is that is scalable and cost effective. Primary mouse fibroblasts are kept in culture for no more than 4 passages to preserve their primary status, and their conditioned medium is then processed by differential centrifugation to obtain crude exosomal preparations that are further purified by sucrose density gradients. This protocol has been successfully used to assess and compare the protein composition of exosomes released by normal mouse fibroblasts and by activated fibroblasts (myofibroblasts) cultured from the GA muscle of WT and Neu1–/– mice, respectively (van de Vlekkert et al., 2019). The Neu1–/– mouse is a model of the lysosomal storage disease sialidosis, caused by deficiency of the sialidase neuraminidase 1 (NEU1) (de Geest et al., 2002; d’Azzo et al., 2015). Mouse cells deficient for Neu1, including primary myofibroblasts, have excessive exocytosis of soluble lysosomal contents as well as increased release of exosomes (van de Vlekkert et al., 2019). Exosomes purified from Neu1–/– myofibroblasts were shown to be potent carriers of fibrotic signaling molecules and to be responsible for eliciting a pathogenic cascade, which ultimately led to muscle fibrosis and degeneration (van de Vlekkert et al., 2019).
Materials and Reagents
Note: All materials and reagents are kept at room temperature unless otherwise described. For the shelf life and storage temperature of reagents, we refer to the manufacturer’s instructions.
- Advantus T-pins (Fisher scientific, catalog number: S174301)
- Light duty paper wipes (Georgia Pacific, Brawny, catalog number: 29221)
- Falcon 50 ml conical centrifuge tubes (Corning 352070) (Fisher Scientific, catalog number: 14-432-22 )
- 10-cm and 15-cm dishes (Midwest Scientific, TPP, catalog numbers: TP93100 and TP93150 respectively)
- Countess cell counting chamber slides and Trypan Blue (Thermo Fisher Scientific, Invitrogen, catalog number: C10228 )
- Thin wall Polypropylene tubes (25 x 89 mm) (Beckman Coulter, catalog number: 326823 )
- Ultra-clear tubes (25 x 89 mm) (Beckman Coulter, catalog number: 344058 )
- Ultra-clear tube (14 x 89 mm) (Beckman Coulter, catalog number: 344059 )
- Premium microcentrifuge tubes (1.7 ml) (Fisher Scientific, Midwest Scientific, catalog number: AVSC1510)
- Multiple well plates (96-well) (Midwest Scientific, TPP, catalog number: TP92696 )
- Protected disposable scalpels (stainless steel blade) (Fisher Scientific, Aspen Surgical Bard-Parker, catalog number: 372610 )
- Immobilon-P Transfer membranes (Millipore, catalog number: IPVH00010 )
- Blotting paper (Fisher, Whatmann, catalog number: 09-301-404)
- Sterile cell strainers (Thermo Fisher Scientific, Fisherbrand, catalog number: 22-363-548 )
- Sterile pipet 5 ml, 10 ml and 25 ml (Falcon, catalog numbers: 357543 , 357551 , 357525 , respectively)
- AvantGuard low-binding barrier tips
10 μl universal-long (Midwest Scientific, catalog number: ASAVANBARTIPS AV10R-H)
20 μl universal (Midwest Scientific, catalog number: ASAVANBARTIPS AV20H)
200 μl universal (Midwest Scientific, catalog number: AV200-H )
1,250 μl universal (Midwest Scientific, catalog number: AV1250H) - Gel round tips (Midwest Scientific, catalog number: MPT1000)
- Adult mice
Notes:- Both male and female mice can be used for this protocol.
- The gastrocnemius muscle is used as example in this protocol. However, the procedure is feasible with any other type of muscle.
- Ethanol 140 proof (Pharmco by Greenfield Global, catalog number: 111000140)
- Ethanol 200 proof (Pharmco by Greenfield Global, catalog number: 111000200 )
- Dulbecco’s Phosphate Buffered Saline (DPBS) (Corning, catalog number: 21-031-CV )
- Collagenase P (100 mg) (Roche, catalog number: 11 213 857 001)
- Dispase II (neutral protease, grade II) (Roche, catalog number: 04 942 078 001)
- CaCl2 (Sigma, catalog number: C1016-100G)
- NaCl (Fisher Scientific, catalog number: S271 )
- Dulbecco’s Modification Eagles Medium (DMEM) (Corning, catalog number: 15-013-CV )
- Dimethyl sulfoxide (DMSO) (Fisher BioReagent, catalog number BP213-100)
- Fetal Bovine Serum (FBS) (Gibco, catalog number: 10437-028 )
- Penicillin-Streptomycin (10,000 U/ml) (Thermo Fisher Scientific, Gibco, catalog number: 15140-122 )
- GlutaMAX supplement (Thermo Fisher Scientific, Gibco, catalog number: 35050-061 )
- Stericup Quick release-GP sterile vacuum filtration system (Millipore, catalog number: S2GPU05RE)
- Triton X-100 solution (10%) (Sigma, catalog number: 93443 )
- TrypLE Express Enzyme (no phenol red) (Thermo Fisher Scientific, Gibco, catalog number: 12604-013 )
- BCA protein assay kit (Thermo Fisher Scientific, Pierce, catalog number: 23225 )
- Criterion TGX stain-free protein gel
10% 18-well, midi-gel (Bio-Rad, catalog number: 5678034)
12% 18-well, midi-gel (Bio-Rad, catalog number: 5678044)
10% 10-well, mini-gel (Bio-Rad, catalog number: 4568033)
12% 10-well, mini-gel (Bio-Rad, catalog number: 4568043) - Precision Plus Protein, all blue pre-stained protein standards (Bio-Rad, catalog number: 1610373 )
- Precision Plus Protein, Unstained protein standards (Bio-Rad, catalog number: 1610363 )
- Tris base (Bio-Rad, catalog number: 1610719 )
- Hydrochloric acid (HCl) (Fisher Scientific, catalog number: A144S-500 )
- Glycine (Bio-Rad, catalog number: 1610718 )
- Sodium dodecyl sulphate (SDS) (Bio-Rad, catalog number: 64086876)
- Laemmli sample buffer (4x) (Bio-Rad, catalog number: 1610747 )
- Dithiothreitol (DTT) (Sigma/Millipore, Roche, catalog number: 10708984001 )
- Methanol (Fisher Scientific, catalog number: A4121)
- Non-fat dry milk (Labscientific, catalog number: M0842)
- Bovine Serum Albumin (BSA) Fraction V (Roche, catalog number: 03116964001)
- Primary and secondary antibodies (Table 1)
Table 1. Primary and secondary antibodies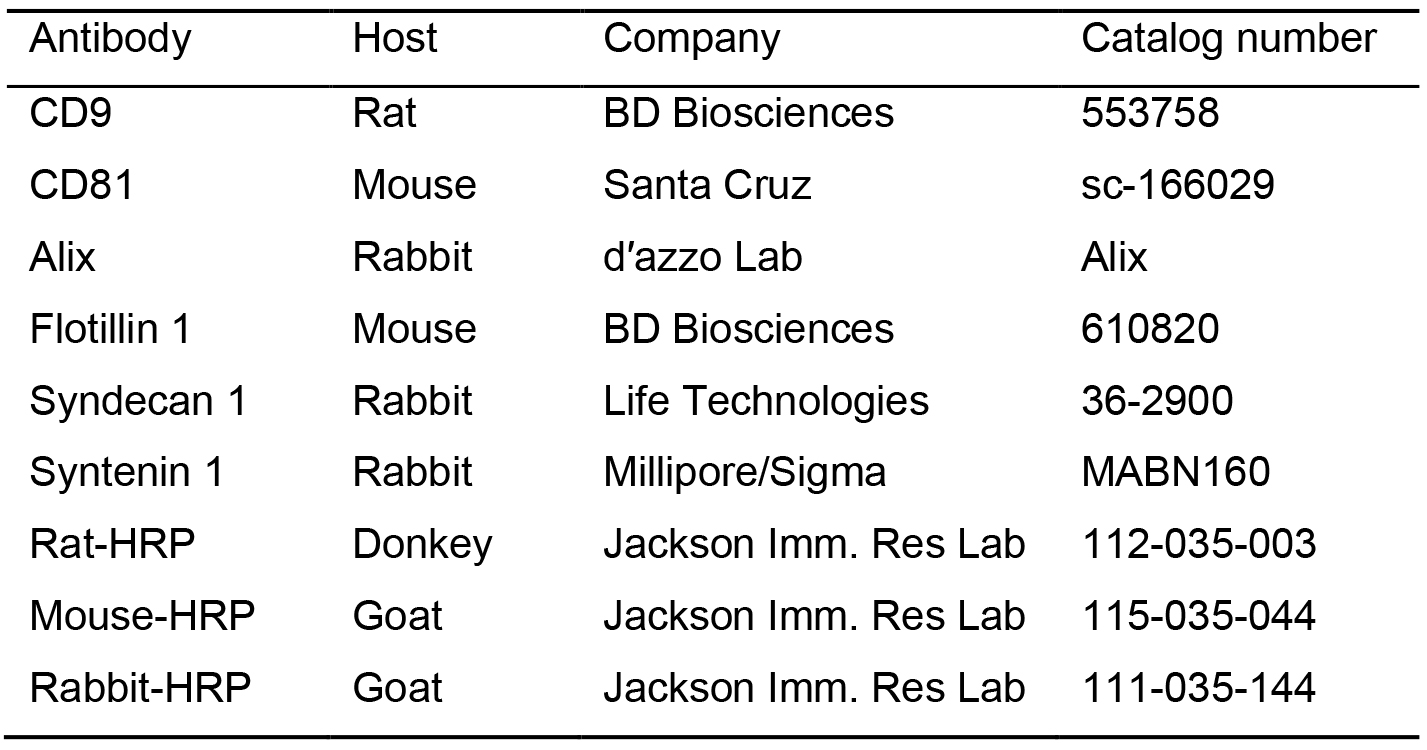
- Sucrose (Fisher Scientific, Fisher Chemical, catalog number: S5-500 )
- cOmplete protease inhibitor cocktail (Millipore/Sigma, Roche, catalog number: 11697498001 )
- 1.0 M Magnesium acetate solution (Mg(Ac)2) (Sigma, catalog number: 63052-100ml)
- Trichloroacetic Acid (TCA) (Sigma, catalog number: 91228-100G)
- Tween-20 (10%) (Bio-Rad, catalog number: 161-0781)
- SuperSignal west Femto (Thermo Fisher Scientific, catalog number: 34096 )
- Collagenase P stock solution (4x) (see Recipes)
- Dispase II stock solution (10x) (see Recipes)
- CaCl2 (1 M) (see Recipes)
- Digestion mixture (see Recipes)
- Complete medium (see Recipes)
- Freezing medium (see Recipes)
- Exosome free FBS (see Recipes)
- Complete exosome free medium (see Recipes)
- 0.1% Triton X-100 (see Recipes)
- BCA reagent working solution (see Recipes)
- 1.0 M DTT (see Recipes)
- 0.5 M Tris-HCl, pH 7.4 (see Recipes)
- Protease inhibitor stock solution (100x) (see Recipes)
- 10 mM Tris-HCl pH 7.4/1 mM Mg(Ac)2 (see Recipes)
- Sucrose gradient working solution (11 ml per sample) (see Recipes)
- 100% TCA (see Recipes)
- 80% ethanol (see Recipes)
- Western blot running buffer (10x) (see Recipes)
- Western blot running buffer (1x) (see Recipes)
- Transfer buffer (10x) (see Recipes)
- Transfer buffer (1x) (see Recipes)
- 20x Tris-buffered saline (TBS) (see Recipes)
- TBS-T(1 L) (see Recipes)
- Blocking buffer (see Recipes)
- 3% BSA-TBS-T (see Recipes)
- SuperSignal west Femto solution (per membrane) (see Recipes)
Equipment
- Aluminum dissection pan with vinyl dissecting pad (Carolina, catalog number: 629004)
- Dissection tools: sterilized scissors and forceps (Roboz, catalog numbers: RS-5983, RS-5877, RS-5358, RS-5135 )
- O2/CO2 incubator (Sanyo, catalog number: MC0-18M)
- Biosafety hood 1300 series A2 (Thermo Scientific, model: 1377)
- Water bath Isotemp 220 (Fisher Scientific, catalog number: FS220)
- Ice bucket (no particular brand is required)
- Centrifuge 5430R with rotor FA-35-6-30 (Eppendorf, catalog number: 022620659) and rotor 5430/R FA-45-48-11 (Eppendorf, catalog number: 5427754008)
- Countess automated cell counter (Thermo Fisher Scientific, Invitrogen, catalog number: C10227 )
- Ultra-centrifuge Optima XPM (Beckman Coulter, catalog number: A99842)
- Alphapette Pipettors
0.5-10 μl (Midwest Scientific, catalog number: ASALPHA A-10)
2-20 μl (Midwest Scientific, catalog number: ASALPHA A-20)
20-200 μl (Midwest Scientific, catalog number: ASALPHA A-200)
100-1,000 μl (Midwest Scientific, catalog number: A-1000 ) - Pipetboy acu 2 (Fisher Scientific, Integra Biosciences Corp, catalog number :155015)
- Criterion vertical electrophoresis cell (midi) (Bio-Rad, catalog number: NC0165100 or 1656001 )
- Mini-Protean tetra vertical electrophoresis cell (Bio-Rad, catalog number: 1658004 )
- PowerPac HC High-current power supply (Bio-Rad, catalog number: 1645052 )
- Chemidoc MP imaging system (Bio-Rad, catalog number: 12003154)
- Mini Trans-Blot electrophoretic transfer cell (Bio-Rad, catalog number: 1703930 )
- Criterion Blotter with plate electrodes (Bio-Rad, catalog number: 1704070 )
- Incubation containers for Western blots; the lids of pipet tip racks
- Tube rack for 50 ml tubes
- Fluostar Omega multi-mode microplate reader (BMG Labtech)
- Plexiglass (in house made)
- Belly dancer shaker (IBI scientific, catalog number: BDRAA115S)
- Stirrer (Fisher Scientific, Isotemp, catalog number: 11-100-49S )
- Stirring bar (Fisher Scientific, catalog number: 50-999-347)
Software
- Image Lab, Version 6.0.1 build 34 (2017) (Bio-Rad)
- Omega, Version 3.00R2 (BMG Labtech)
- Omega Analysis, Version 2.40, Build 13 (BMG Labtech)
Procedure
- Muscle excision
- Euthanize the mouse following guidelines approved by the institution.
Notes:- Both male and female mice can be used.
- For this protocol a genetically engineered animal model of the lysosomal storage disease sialidosis (de Geest et al., 2002) crossed into C57Bl/6 or FVB background was used.
- The isolation of the hind limb muscles does not require the use of a biosafety cabinet. No sterile conditions are necessary.
- Pin the mouse down in a prone position (dorsal side up) to the dissecting pad (Figure 1A). Spray the mouse with 70% ethanol.
Note: For the isolation of other muscle types, pin the mouse down in a supine position (ventral side up) to the dissecting board. - Remove the skin from both hind limbs to expose the muscles and gently cut away the fascia of both hind limb muscles. Locate the Achilles tendon and separate it from the bone with a pair of forceps (Figure 1B).
Notes:- Shaving the fur is not necessary for this procedure.
- There is not a standard method for the removal of the fur/skin to expose the muscles. The procedure we use is as follows: Make a ~1 cm cut in the skin at the level of the hip joint and carefully pull the skin towards the ankle to expose the leg muscles.
- Cut the Achilles tendon and carefully pull the GA muscle upwards towards the knee joint and cut it loose at the tendon (Figures 1C and 1D).
Note: As the tendons from the GA and soleus are interconnected (forming the Achilles tendon) it is not possible to separate them. - Remove the soleus (darker red colored part of the muscle). When the GA and soleus muscles are cut at the knee joint, the soleus will detach from the GA muscle and can be lifted up and cut at the Achilles tendon to separate it from the GA muscle (Figures 1E and 1F).
Note: The removal of the soleus allows for a purer fibroblast isolation from the remaining GA muscle. - Weight a 50 ml tube containing 5 ml DPBS.
- Combine the GA muscles from both legs and transfer them to the pre-weighted 50 ml tube and weight the tube again. Place the tube on ice.

Figure 1. Dissection of GA muscle from the hind limb of adult mice. A. Picture presented here shows the mouse legs before removal of the fur/skin. B. The fur/skin is removed to expose the GA muscle (dashed lines) and the Achilles tendon, which is separated from the bone by a pair of forceps. C. The Achilles tendon is cut. D. The GA/soleus muscles are pulled upwards and cut at the knee joint. E. Isolated GA muscle is shown with the soleus (darker red and dashed lines) that is still attached. F. GA muscle is shown after removal of the soleus.
- Euthanize the mouse following guidelines approved by the institution.
- Fibroblasts isolation from the gastrocnemius muscle
- Prepare the biosafety hood by placing all necessary tools inside, including tube racks, pipets, dishes, tubes, tip boxes.
- Take two scalpels out of the sterile wrap and place them in the hood.
- Place a 10 cm dish open in the hood.
- Take out the sterile forceps and transfer the two GA muscles into the 10 cm dish.
Note: The tissues contain residual amount of DPBS, which keeps the tissue moist during the procedure. - Chop the muscles with the scalpels until it become a fine paste.
- Transfer the finely chopped muscle paste onto a 50 ml tube using the scalpels, cap the tube and place it back on ice.
Notes:- Additional DPBS is not required to transfer the minced muscle tissue with scalpels.
- If additional muscle samples need to be processed, repeat steps B4-B6 and transfer each chopped muscle paste to a new 50 ml tube.
- Add digestion mixture to each tube; 3.5x volume/mg tissue (see Recipes 1-4) and incubate at 37 °C for 45 min.
Note: Alternatively, a tissue dissociator can be used to dissociate the muscles. - During the 45 min digestion period, the tissue suspension needs to be mixed every 10 min by pipetting it up and down with a 5 ml pipet. This will promote the complete dissociation of the tissue.
- Add 20 ml complete medium to each tube to terminate the enzymatic digestion (see Recipe 5).
- Place a 70 μm cell strainer on a new 50 ml tube.
- Filter the tissue/cell suspension through the strainer, rinsing it with an additional 5.0 ml complete medium.
- Centrifuge the tissue/cell suspension at 300 x g at 4 °C for 10 min.
- Resuspend the pellet containing the cells in 10 ml complete medium.
- Seed the resuspended pellet in a 10 cm dish (Passage 0, P0) and incubate at 37 °C/5% CO2/3% O2.
Note: The fibroblasts are cultured at a low oxygen level to keep the culture conditions as close as possible to the physiologic conditions, since the O2 level in skeletal muscle is around 2.5% (Mas-Bargues et al., 2019).
- Fibroblasts culture, trypsinization and expansion
- Culture the P0 fibroblasts until 100% confluency plus 1 day.
Notes:- Primary fibroblasts at P0 are routinely cultured until 100% confluency plus 1 day (and only for P0) to purge the cultures from any contaminating cells.
- The expected yield of fibroblasts (P0) from one pair of hind limb gastrocnemius muscles ranges from 1 to 5 x 106 cells, depending on the age and background of the mouse used.
- When cells are ready to be expanded, remove the culture medium and rinse the cells with 10 ml DPBS.
Note: Be sure to remove all DPBS because it may interfere with the TrypLE express enzymatic activity. - Add 1.0 ml TrypLE to the dish and incubate at 37 °C for 5 min.
- Tap the side of the dish and check under the microscope for complete detachment of the cells. If necessary, incubate another 5 min at 37 °C.
- Add 10 ml complete medium to inactivate the TrypLE and transfer the cells into a 50 ml tube.
- Centrifuge at 300 x g for 10 min at room temperature.
Note: At this point P0 cells can be resuspended in freezing medium to be frozen for later use (see Recipe 6). - Resuspend the cells in 10 ml complete medium.
- If needed, cells can be counted by mixing 10 μl cell suspension with 10 μl Trypan Blue and transferred to the Countess cell counter chamber.
Note: Any type of cell counter can be used as long as it gives an accurate count of viable cells. - Cells are seeded at a concentration of 1.0 x 105/ml, resulting in 2 x 106 cells (20 ml) per 15 cm dish.
- Culture the cells at 37 °C/5% CO2/3% O2 until 80%-90% confluency (Passage 1, P1).
- Repeat Steps C2 to C10 for additional passaging of the culture.
Notes:- The number of 15 cm dishes needed will depend on the total number of cells required; e.g., at P2 cells are seeded in 2-4, 15 cm dishes; at P4 they are seeded in 15-30, 15 cm dishes if a high yield of exosomes is required.
- The yield of exosomes from a 15 cm dish of primary fibroblasts is ~3-4 μg. We suggest seeding 20-25, 15 cm dishes to obtain enough exosomes for subsequent analyses.
- After P4, primary cells are discarded and not used for any further experiments as they change their characteristics.
- Culture the P0 fibroblasts until 100% confluency plus 1 day.
- Crude Exosome isolation
- Wash the 15 cm dishes containing fibroblasts (P2-P4) with 10 ml DPBS.
Note: Be sure to remove all DPBS because it may interfere with the TrypLE express enzymatic activity. - Add 2.5 ml TrypLE to each dish and incubate at 37 °C for 5 min.
- Tap the side of the dish and check under the microscope for complete detachment of the cells. If necessary, incubate another 5 min at 37 °C.
- Add 10 ml complete medium to inactivate the TrypLE and transfer the cells to a 50 ml tube.
- Centrifuge the cells at 300 x g for 10 min at room temperature.
- Resuspend the cells in 10 ml complete exosome free medium (see Recipes 7 and 8).
Note: All vesicles/exosomes naturally present in FBS need to be removed prior to adding the FBS to the fibroblast culture medium, as they will interfere with further experimental analyses. - If needed, cells can be counted by mixing 10 μl cell suspension with 10 μl Trypan Blue and transferred to the Countess cell counter chamber.
Note: Any type of cell counter can be used as long as it gives an accurate count of viable cells. - Seed the cells in the exosome free medium at the concentration of 1.5 x 106 - 2.0 x 106 per 15 cm dish and incubate at 37 °C/5% CO2/3% O2.
- Collect the conditioned medium in 50 ml tubes on ice after 16 to 24 h.
Notes:- The collected medium can be stored at 4 °C up to one week, if not processed immediately.
- If cells are not 80% confluent at the time of medium harvesting, fresh exosomal medium can be added and collected after an additional 16 to 24 h period. Combine the two batches of conditioned medium for further processing.
- Centrifuge the conditioned medium at 300 x g for 10 min at 4 °C and transfer it to a new 50 ml tube on ice (Figure 2.1).
Note: This step is performed to eliminate viable cells. - Centrifuge at 2,000 x g for 10 min at 4 °C (Figure 2.2).
Note: This step is performed to eliminate dead cells. - Transfer supernatant to a thin wall Polypropylene tube on ice and centrifuge in an ultracentrifuge at 10,000 x g at 4 °C for 30 min (Figure 2.3).
Note: This step removes organelles, apoptotic bodies and membrane fragments. Centrifugation at 10,000 x g can also be performed using a Sorval centrifuge. - Transfer the supernatant to an ultra-clear tube on ice and ultracentrifuge it at 100,000 x g at 4 °C for 1.5 h (Figure 2.4).
Note: If exosomes need to be under sterile condition, the open ultra-clear tubes can be exchanged for sterile ultra-clear tubes with lids. - Carefully discard the supernatant with a 10 ml pipette. Leave about 1.0 ml of liquid in the tube to avoid disturbance of the pellet.
- Add an additional 1.0 ml of ice-cold DPBS and resuspend the pellet using a P1000 pipette.
- Fill the tube up to 35 ml with ice-cold DPBS and ultracentrifuge at 100,000 x g at 4 °C for 1.5 h (Figure 2.5).
- Carefully discard the supernatant and leave the tube containing the pellet on ice.
Note: The pellet should be translucent and may be hard to visualize, therefore the supernatant should be removed very carefully. - Resuspend the pellet in an appropriate volume of ice-cold DPBS (~25 μl/15 cm dish).
- The exosomes can be kept at 4 °C up to one week. For long-term storage exosomes should be stored at -80 °C.
Notes:- Avoid freezing/thawing cycles, which can damage exosomes and reduce their numbers. The exosomes should be aliquoted if multiple experiments are performed.
- The crude exosomes can be used for a variety of analyses, including Western blots and in vitro uptake experiments.
- When needed, mass spec analysis can be performed on crude exosomes. Run fifty μg of crude exosomes on a 10% bis-tris gel for ~1 cm into the gel and cut 3 equal sized bands for in-gel digestion (sequencing grade modified trypsin solution) prior to LC-MS/MS.
- Wash the 15 cm dishes containing fibroblasts (P2-P4) with 10 ml DPBS.
- Quantification of crude exosomes for Western blot analysis
Note: As an alternative, nanoparticle tracking analysis (NTA) as well as dynamic light scattering (DSL) can be used to characterize exosomes (Szatanek et al., 2017).- Pipet 10 μl of each albumin standard dilutions (see Table 2) in duplicate into a 96-well plate to obtain the BCA standard curve.
Table 2. BCA Standard Curve preparation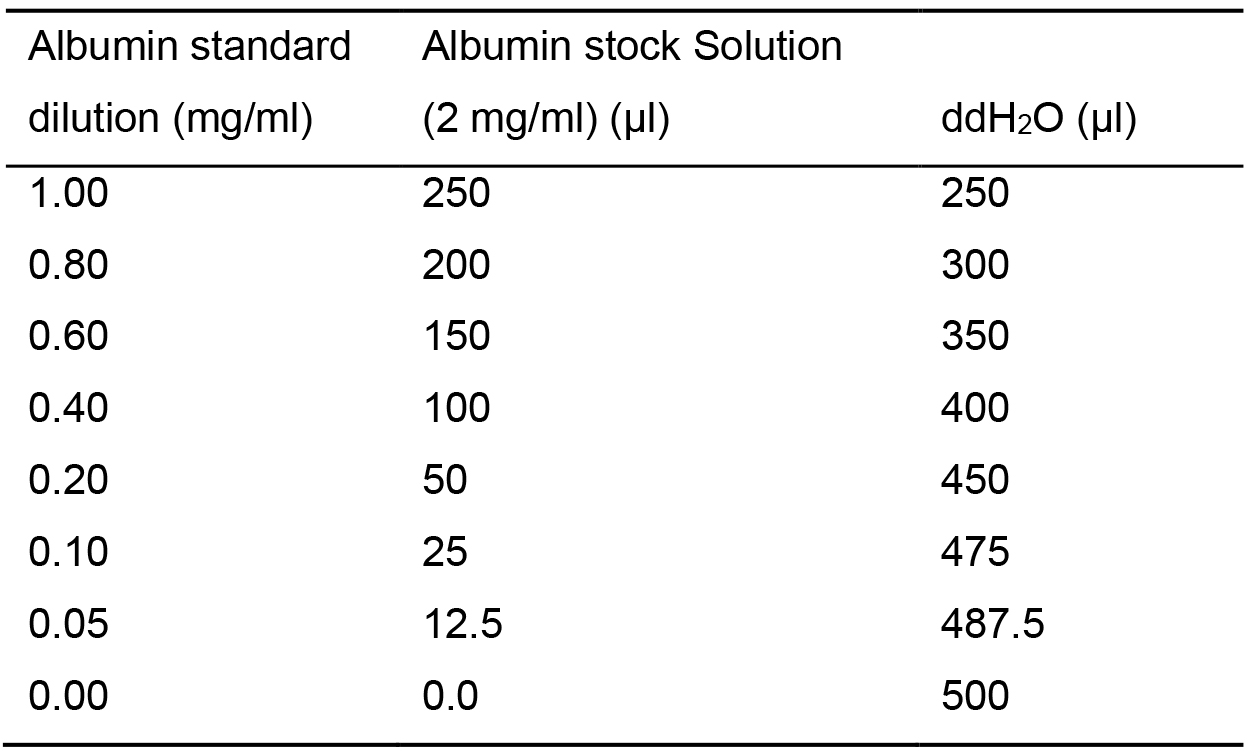
- Pipet 5.0 μl 0.1% Triton X-100 in ddH2O in duplicate in a 96-welll plate (see Recipe 9).
- Add 5.0 μl of the exosomal samples to the wells containing 0.1% Triton X-100, resulting in a 1:2 dilution.
Note: The resuspended exosomal pellet should be diluted further (1:5 or 1:10), depending on the final yield of the preparation. - Add 200 μl BCA working solution to all wells (see Recipe 10).
- Incubate the plate at 37 °C for 30 min and read the protein concentration at OD562 using a plate reader.
- On average 1 x 106 primary fibroblasts will yield approximately 3-4 μg exosomes that can be resuspended directly into electrophoresis loading buffer (0.5 μl of 1.0 M DTT (Recipe 11), 5.0 μl Laemmli sample buffer (4x) and 14.5 μl of ddH2O).
- Pipet 10 μl of each albumin standard dilutions (see Table 2) in duplicate into a 96-well plate to obtain the BCA standard curve.
- Sucrose density gradient for purification of exosomes
- Prepare all the solutions needed for the sucrose density gradient and keep them on ice (see Recipes 12-15 and Table 3).
Table 3. Sucrose gradient solutions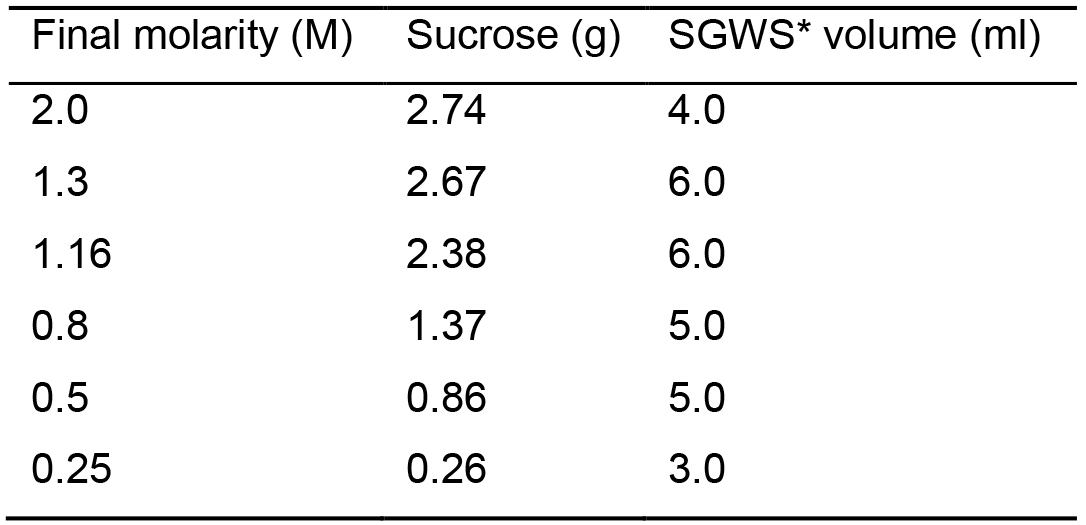
*SGWS: Sucrose Gradient Working Solution (see Recipe 13). - Resuspend 30 to 100 μg of exosomes in 1.0 ml of 0.25 M sucrose solution.
- Prepare the gradient in a 13 ml ultra-clear tube by pipetting the sucrose solutions in sequence starting from the bottom of the tube with 1.5 ml of 2.0 M, 2.5 ml of 1.3 M, 2.5 ml of 1.16 M, 2.0 ml of 0.8 M, 2.0 ml of 0.5 M sucrose. Layer the gradient at the top with 1.0 ml sucrose solution containing exosomes (Figure 2.6).
- Centrifuge the gradient at 100,000 x g at 4 °C for 2.5 h, using a swinging bucket rotor (Figure 2.6).
- Carefully take the tubes out of the rotor and place them on ice.
- Collect 1.0 ml fractions starting from the top of the gradient and transfer them to 1.7 ml Eppendorf tubes on ice (Figure 2.7).
- Add 110 μl 100% TCA to each tube, mix well and incubate at room temperature for 10 min (see Recipe 16) (Figure 2.8).
- Centrifuge at 10,000 x g at room temperature for 10 min and discard the supernatant.
- Wash the pellets with 500 μl pre-cooled 80% ethanol, pipetting up and down, and place the resuspended pellets at -20 °C for 10 min (see Recipe 17).
- Centrifuge at 10,000 x g at 4 °C for 10 min.
- Remove carefully the supernatant and let the pellets air dry for 10-15 min.
- Resuspend the pellets in 15-30 μl ice cold DPBS.
Notes:- The volume of DPBS used to resuspend the pellets depends on the starting material and the size of the pellet.
- For Western blot analyses, resuspend each pellet in electrophoresis loading buffer (0.5 μl 1.0 M DTT (Recipe 16), 5.0 μl Laemmli sample buffer (4x) and 14.5 μl of ddH2O).
- Measure the protein concentration according to Steps E1 to E5.

Figure 2. Schematic presentation of exosomes isolation from cultured media. Removal of cells (1), dead cells (2) and cells debris (3) after sequential centrifugation steps. Ultra-centrifugation of exosomes (4) and washing of exosomal pellet in DPBS (5). Loading of exosomal pellet on a sucrose density gradient (6). Fraction collection (7) after ultra-centrifugation and precipitation with TCA (8).
- Prepare all the solutions needed for the sucrose density gradient and keep them on ice (see Recipes 12-15 and Table 3).
- Western blot analyses of crude exosomes and purified sucrose gradient samples
- Prepare the electrophoresis running buffer and the buffer for transferring the proteins from the gel onto a PVDF membrane (Recipes 18-21).
- For the protein transfer, cut filter papers and membranes according to the size the gels used.
Note: Filter papers and PVDF membranes are cut 7 cm by 9 cm for mini gels and 9 cm by 14 cm for midi gels. - Assemble the precast gel in the electrophoresis tank and fill the tank with running buffer.
Notes:- See instruction manual from Bio-Rad (https://www.bio-rad.com/) for “Mini-PROTEAN Tetra Cell Precast”.
- Gels with different percentage of polyacryamide are used depending on the molecular weight of the proteins that need to be resolved: 7.5% for proteins of 50-200 kDa, 10% for proteins of 15-100 kDa, 12% for proteins of 10-70 kDa and 15% for proteins of 12-45 kDa.
- Boil the exosomes (from crude exosomal preparations or purified sucrose density gradient fractions) resuspended in loading buffer for 5 min. Spin the samples at ~10,000 x g for 30 s prior to loading them on a precast gel.
- Before loading of the samples, rinse the wells of the gel with running buffer.
- Load the precast gel with the exosomal samples and the protein markers, using round loading tips.
Note: Resuspend each sample thoroughly prior to loading the gel. - Run the gel at 100 V until the dye of the loading buffer reaches the base of the gel.
- Carefully remove the gel from the cassette, placed it on the Chemidoc imaging tray containing 1.0 ml of transfer buffer and image it, using a Stain-Free program (Figure 3).
Note: The stain-free gel is a handy tool to assess if the loading is equal in all samples.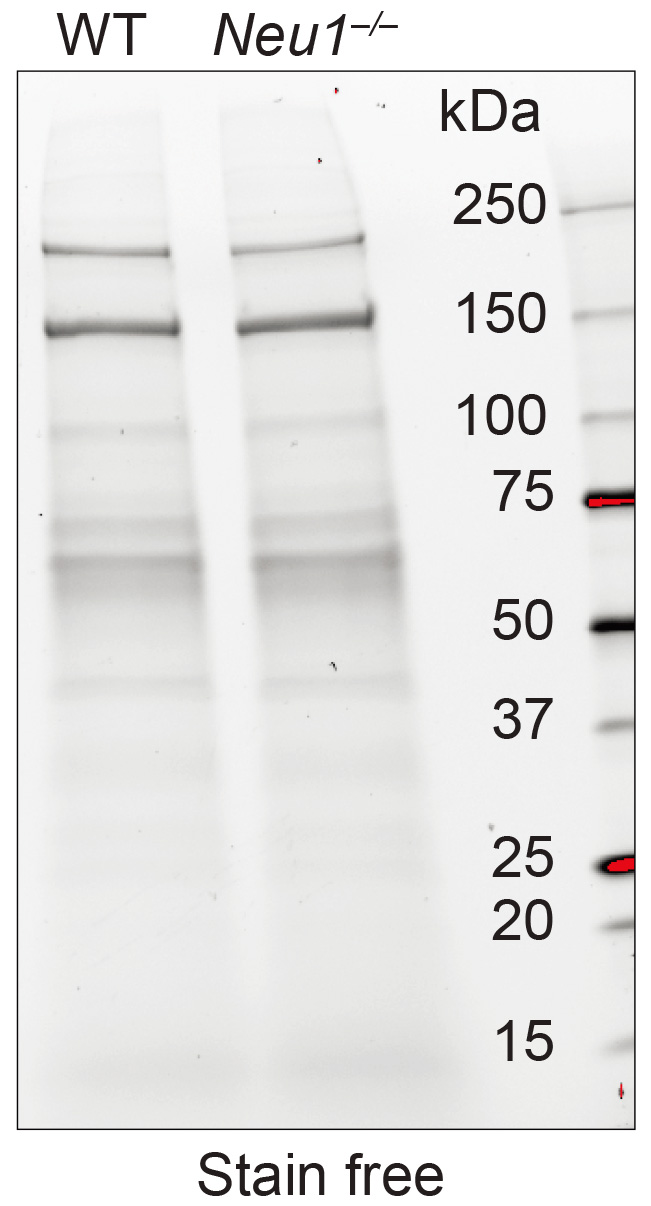
Figure 3. Stain-free gel loaded with exosomes. WT and Neu1–/– crude exosomes run on a Criterion TGX Stain-Free precast gel and imaged using the chemidoc stain free program (Figure from van de Vlekkert et al., 2019). - Equilibrate the gel in transfer buffer.
- Label the PVDF membrane with a pencil or pen and activate it in 100% methanol for 10 s (max 30 s).
- Briefly rinse the PVDF membrane in ddH2O and equilibrate it in transfer buffer.
- Soak the filter papers in transfer buffer.
- Assemble the gel, filter papers and PVDF membrane for transfer.
Note: See instruction manual from Bio-Rad (https://www.bio-rad.com/) for “Mini Trans-Blot Electrophoretic Transfer Cell”. - Transfer at 80 V for 1.5 to 2 h or overnight at 30 V at 4 °C.
- After transfer incubate the membrane in blocking buffer at room temperature for 1 h with agitation (see Recipes 22-24).
- Incubate the membrane (in a 50 ml tube) with the primary antibody (see Table 1) diluted in 3.0 ml 3% BSA-TBS-T overnight at 4 °C with gentle agitation (see Recipes 22-23, 25).
Note: Dilute the primary antibody to the recommended concentration/dilution. - Rinse the membrane 1 time with TBS-T and wash 3 times 10 min with TBS-T under gentle agitation (see Recipes 22-23).
- Incubate 1 h with a secondary antibody (see Table 1; 1:10,000 dilution) in blocking buffer at room temperature under gentle agitation.
- Rinse the membrane 1 time with TBS-T and wash 3 times 10 min with TBS-T under gentle agitation.
- Pipet 1.0 ml of SuperSignal west Femto developer solution on a plexiglass plate, making sure that the table/bench is leveled to avoid run off of the solution (Recipe 26).
- Remove excess of TBS-T and place the membrane face down on 1.0 ml developer solution.
- Incubate 30 s to 1 min, and place the membrane face up on the Blot/UV/Stain-Free sample tray.
- Image the membranes (Figure 4).
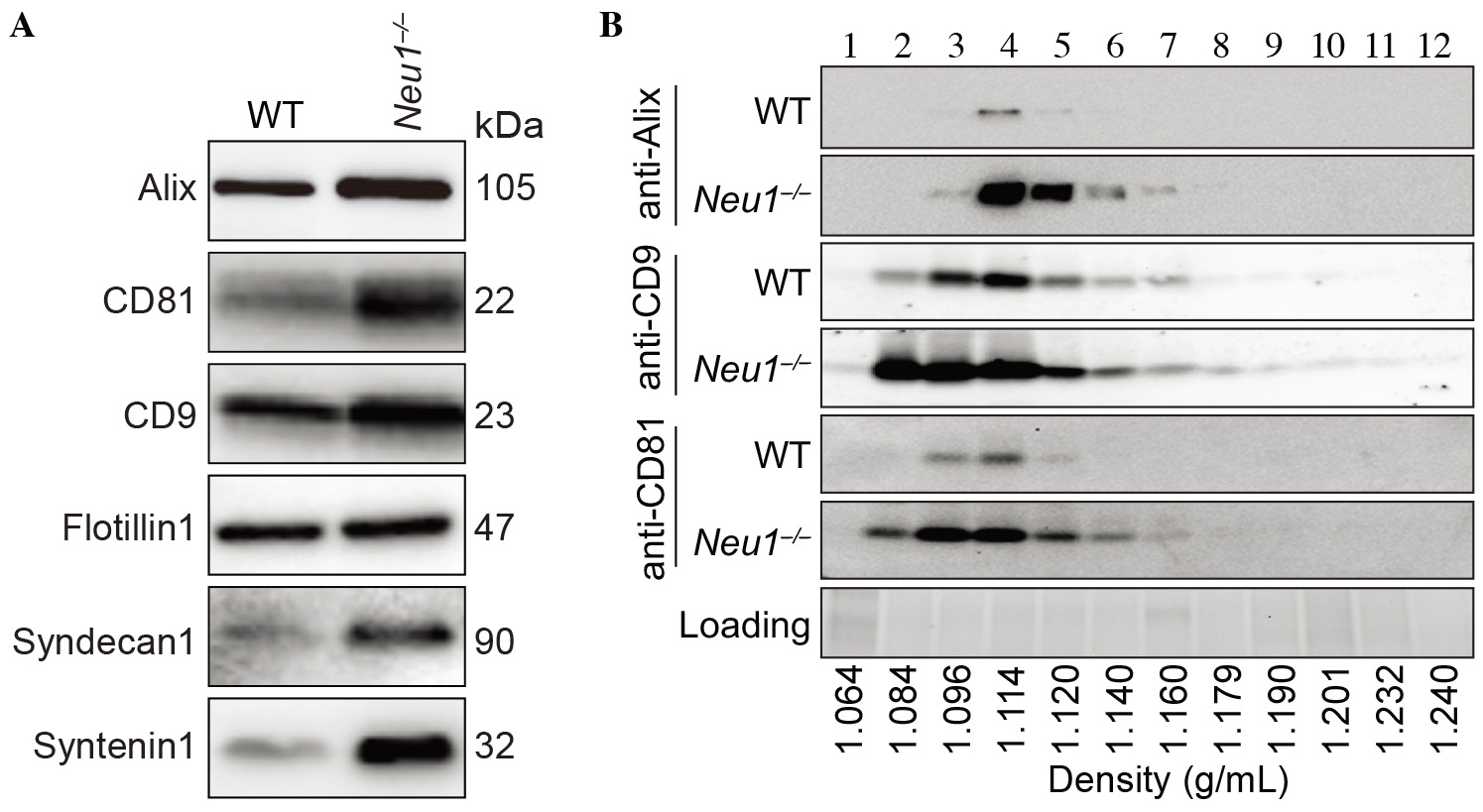
Figure 4. Immunoblot analyses of exosomes using canonical exosomal markers. A. Exosomes were purified from WT and Neu1–/– cultured (myo)fibroblasts and probed with anti-Alix, CD81, CD9, flotillin1, syndecan1 and syntenin1 antibodies. B. WT and Neu1–/– crude exosomes were subjected to sucrose density gradient and probed with anti-Alix, CD9 and CD81 antibodies. Exosomes sedimented in fractions 3-6 which correspond to the densities of 1.096-1.140 g ml-1 (figure from van de Vlekkert et al., 2019).
Data analysis
- The protein concentration is calculated using the Omega data analysis software.
- Level of exosomal proteins are quantified using Image Lab software and normalized against the total amounts of proteins loaded on a stain-free gel.
Recipes
- Collagenase P stock solution (4x)
Reconstitute 100 mg in 5.0 ml DPBS (20 mg/ml)
Store at -20 °C - Dispase II stock solution (10x)
Reconstitute 1.0 g in 100 ml DPBS (11 U/ml) and store at -20 °C - CaCl2 (1 M)
Dissolve 1.1098 g in 10 ml ddH2O - Digestion mixture
2.5 ml of Collagenase P stock solution (final conc. 5.0 mg/ml)
2.0 ml of Dispase II stock solution (final conc. 1.2 U/ml)
100 μl of CaCl2 stock solution (final conc. 5.0 mM)
15.4 ml DPBS
Filter sterilize the solution and store at -20 °C - Complete medium
440 ml DMEM
50 ml FBS
5 ml Penicillin-Streptomycin
5 ml GlutaMAX supplement
Filter sterilize the solution and store at 4 °C - Freezing medium
10% DMSO
90% FBS - Exosome free FBS
Spin FBS overnight at 100,000 x g at 4 °C in sterile thin wall Polypropylene tubes and transfer supernatant to 50 ml tubes
Store at -20 °C - Complete exosome free medium
440 ml DMEM
50 ml Exosome free FBS
5 ml Penicillin-Streptomycin
5 ml GlutaMAX supplement
Filter sterilize the solution and store at 4 °C - 0.1% Triton X-100
Mix 1.0 ml Triton X-100 (10%) with 99 ml ddH2O - BCA reagent working solution
Stable for 1 day
Mix 50 parts of BCA reagent A with 1 part of reagent B - 1.0 M DTT
10 g DTT
64.83 ml ddH2O
Make 1.0 ml aliquots and store at -20 °C - 0.5 M Tris-HCl, pH 7.4
6.05 g Tris-base
95 ml ddH2O
Adjust pH to 7.4 with HCl
Finalize volume to 100 ml and autoclave - Protease inhibitor stock solution (100x)
Dissolve one cOmplete protease inhibitor cocktail tablet in 500 μl ddH2O
Store at -20 °C - 10 mM Tris-HCl pH7.4/1 mM Mg(Ac)2
100 μl Mg(Ac)2 of 1.0 M solution
2.0 ml of 0.5 M Tris-HCl pH 7.4
97.9 ml ddH2O
Store at 4 °C - Sucrose gradient solution
30 ml 10 mM Tris-HCl pH7.4/1 mM Mg(Ac)2
300 μl protease inhibitor (100x)
Keep the solution on ice until use - 100% TCA
100 g TCA powder in 100 ml ddH2O - 80% ethanol
80 ml 100% ethanol
20 ml ddH2O - Western blot running buffer (10x)
30.28 g Tris Base
144.13 g Glycine
10.0 g SDS
800 ml ddH2O
Check pH and it should be 8.3
Finalize volume to 1.0 L - Western blot running buffer (1x)
Prepare fresh before use
100 ml Western blot running buffer (10x)
900 ml ddH2O - Transfer buffer (10x)
30.28 g Tris Base
144.13 g Glycine
800 ml ddH2O
Finalize volume to 1.0 L with ddH2O - Transfer buffer (1x)
100 ml transfer buffer (10X)
200 ml methanol
700 ml ddH2O
Prepare fresh and keep at 4 °C until use - 20x Tris-buffered saline (TBS)
121.14 g Tris base
175.32 g NaCl
900 ml ddH2O
Adjust pH to 8.0 with HCl
Finalize volume to 1.0 L with ddH2O - TBS-T (1 L)
50 ml 20x Tris-buffered saline (TBS)
10 ml Tween-20 (10%)
940 ml ddH2O to final volume of 1 L and mix well - Blocking buffer
5.0 g Non-fat dry milk
100 ml TBS-T
Store at 4 °C - 3% BSA-TBS-T
3.0 g BSA
100 ml TBS-T
Store at 4 °C - SuperSignal west Femto solution (per membrane)
Mix 0.5 ml Luminol/Enhance with 0.5 ml Stable Peroxide Buffer
Acknowledgments
Funding: This work was supported in part by NIH grants R01GM104981, RO1DK095169, and CA021764, the Assisi Foundation of Memphis, and the American Lebanese Syrian Associated Charities. The protocol was adapted from van de Vlekkert et al., 2019. The authors have no conflict of interest or competing interests to declare.
Ethics
Animals were housed in a fully AAALAC (Assessment and Accreditation of Laboratory Animal Care)–accredited animal facility with controlled temperature (22 °C), humidity, and lighting (alternating 12-h light/dark cycles). Food and water were provided ad libitum. All procedures involving mice were performed according to animal protocols approved by the St. Jude Children’s Research Hospital Institutional Animal Care and Use Committee and according National Institutes of Health guidelines (Animal protocol #235-100465-03/17; exp. 03/01/2017-03/01/2020).
References
- d'Azzo, A., Machado, E. and Annunziata, I. (2015). Pathogenesis, Emerging therapeutic targets and Treatment in Sialidosis. Expert Opin Orphan Drugs 3(5): 491-504.
- de Geest, N., Bonten, E., Mann, L., de Sousa-Hitzler, J., Hahn, C. and d'Azzo, A. (2002). Systemic and neurologic abnormalities distinguish the lysosomal disorders sialidosis and galactosialidosis in mice. Hum Mol Genet 11(12): 1455-1464.
- Gurunathan, S., Kang, M. H., Jeyaraj, M., Qasim, M. and Kim, J. H. (2019). Review of the isolation, characterization, biological function, and multifarious therapeutic approaches of exosomes. Cells 8(4): E307.
- Mas-Bargues, C., Sanz-Ros, J., Román-Domínguez, A., Inglés, M., Gimeno-Mallench, L., El Alami, M., Viña-Almunia, J., Gambini, J., Viña, J. and Borrás, C. (2019). Relevance of oxygen concentration in stem cell culture for regenerative medicine. Int J Mol Sci 20(5): E1195.
- Rackov, G., Garcia-Romero, N., Esteban-Rubio, S., Carrión-Navarro, J., Belda-Iniesta, C. and Ayuso-Sacido, A. (2018). Vesicle-mediated control of cell function: the role of extracellular matrix and microenvironment. Front Physiol 9: 651.
- Szatanek, R., Baj-Krzyworzeka, M., Zimoch, J., Lekka, M, Siedlar, M., and Baran J. (2017). The Methods of Choice for Extracellular Vesicles (EVs) Characterization. Int J Mol Sci 18: 1153.
- van de Vlekkert, D., Demmers, J., Nguyen, X. X., Campos, Y., Machado, E., Annunziata, I., Hu, H., Gomero, E., Qiu, X., Bongiovanni, A., Feghali-Bostwick, C. A. and d'Azzo, A. (2019). Excessive exosome release is the pathogenic pathway linking a lysosomal deficiency to generalized fibrosis. Sci Adv 5(7): eaav3270.
Article Information
Copyright
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
van de Vlekkert, D., Qiu, X., Annunziata, I. and d'Azzo, A. (2020). Isolation, Purification and Characterization of Exosomes from Fibroblast Cultures of Skeletal Muscle. Bio-protocol 10(7): e3576. DOI: 10.21769/BioProtoc.3576.
Category
Cell Biology > Organelle isolation > Exosomes
Cell Biology > Cell isolation and culture > Monolayer culture
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








