- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Preparation of a Bacteriophage T4-based Prokaryotic-eukaryotic Hybrid Viral Vector for Delivery of Large Cargos of Genes and Proteins into Human Cells
Published: Vol 10, Iss 7, Apr 5, 2020 DOI: 10.21769/BioProtoc.3573 Views: 5630
Reviewed by: Miao HeVasudevan AchuthanRakesh Bam

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Assembly and Mutagenesis of Human Coronavirus OC43 Genomes in Yeast via Transformation-Associated Recombination
Brett A. Duguay and Craig McCormick
Aug 20, 2025 3036 Views

Preparation and Characterization of Lipid Nanoparticles Co-loaded With DNA and Nitro-Oleic Acid
Manthan N. Patel [...] Jacob S. Brenner
Sep 20, 2025 3159 Views

Cost-Effective and Reproducible Preparation of mRNA-Loaded Lipid Nanoparticles Using a Conventional Laboratory-Scale Microfluidic Assembly System
Yunqi Li [...] Ruoyang Zhao
Sep 20, 2025 1447 Views
Abstract
A viral vector that can safely and efficiently deliver large and diverse molecular cargos into cells is the holy grail of curing many human diseases. Adeno-associated virus (AAV) has been extensively used but has a very small capacity. The prokaryotic virus T4 has a large capacity but lacks natural mechanisms to enter mammalian cells. Here, we created a hybrid vector by combining T4 and AAV into one nanoparticle that possesses the advantages of both. The small 25 nm AAV particles are attached to the large 120 nm x 86 nm T4 head through avidin-biotin cross-bridges using the phage decoration proteins Soc (small outer capsid protein) and Hoc (highly antigenic outer capsid protein). AAV thus “piggy-backed” on T4 capsid, by virtue of its natural ability to enter many types of human cells efficiently acts as a “driver” to deliver large cargos associated with the T4 head. This unique T4-AAV hybrid vector approach could pave the way for the development of novel therapeutics in the future.
Keywords: Bacteriophage T4Background
There has been a desperate need for new and efficient delivery vehicles that can transport large cargos of genes and proteins into human cells to stimulate the production of therapeutic biomolecules and/or to repair cellular and genetic defects. Such vehicles will allow translation of the rapidly emerging technologies such as CRISPR, CAR T cells and so on, into therapies, both for mass applications as well as for personalized medicine (Stewart et al., 2016).
Assembling nanoparticles with different properties into a hybrid complex is a powerful strategy for developing novel functional materials, as these hybrid complexes exhibit collective and collaborative attributes, some of which might be different from those exhibited by the individual particles (Ghosh et al., 2012; Wang et al., 2014). Virus nanoparticles representing a fascinating class of protein-polynucleotide supramolecular structures are an ideal natural resource to construct such hybrids. Considering different shapes and sizes ranging from tens to hundreds of nanometers and their documented ability for intracellular delivery, combining different viral building blocks into a multifunctional nanoparticle will greatly accelerate future therapies.
Molecular and genetic analyses led to the engineering of an empty phage T4 shell (head or capsid) lacking the outer capsid proteins Soc (small outer capsid protein; 870 copies/head; 9 kDa) and Hoc (highly antigenic outer capsid protein; 155 copies/head; 41 kDa), neck, or tail as a delivery vehicle (Zhang et al., 2011). Both Soc and Hoc are non-essential for phage infection and exhibit high binding affinity and specificity to T4 capsid surface. Soc binds as a trimer at the quasi three-fold axes whereas Hoc binds as a monomer at the center of each gp23 capsomer (hexamer). Foreign proteins can be efficiently displayed on T4 head through fusion to N and C termini of Soc or the N terminus of Hoc which are well exposed (Tao et al., 2013).
The 25 nm adeno-associated virus (AAV) has been extensively used as a gene delivery vehicle but it has a very small capacity and cannot deliver proteins. The 120 x 86 nm prokaryotic virus, the bacteriophage T4, on the other hand has a large capacity but it lacks natural mechanisms to enter human cells. It has no tropism to human cells, has no known toxicity or pathogenicity, and exhibits no preexisting immunity. We reported the development of the phage-virus hybrid vector by integrating these two most well-characterized delivery vehicles into one (Figure 1) (Zhu et al., 2019). A specific biomolecular avidin-biotin interaction was selected as a driving force to mediate the interaction between the T4 head and AAV particle. It is also one of the strongest no-covalent interactions known in nature (Kd = 10-15 M), making it ideal as a molecular bridge between the two nanoparticles (Jain and Cheng, 2017). We engineered it in such a way that all the different compartments of the T4-AAV nanoparticle could be utilized for loading cargo molecules. These include the interior of the T4 capsid shell that can accommodate 170 kbp of DNA cargo (Zhang et al., 2011), the interiors of attached AAV shells each accommodating 5 kbp of DNA cargo (Muzyczka, 2014), and the exterior of the T4 shell that can be decorated with up to 1,025 molecules of protein cargo (Li et al., 2007; Shivachandra et al., 2007; Tao et al., 2013 and 2018). This system has been optimized such that varieties of customized cargos can be prepared by simple mixing of interacting components that have nanomolar or sub-nanomolar affinities and by using a powerful DNA packaging motor (Leffers and Rao, 1996; Fuller et al., 2007). Furthermore, different cargo molecules can be mixed and matched, quantified, and tuned as desired.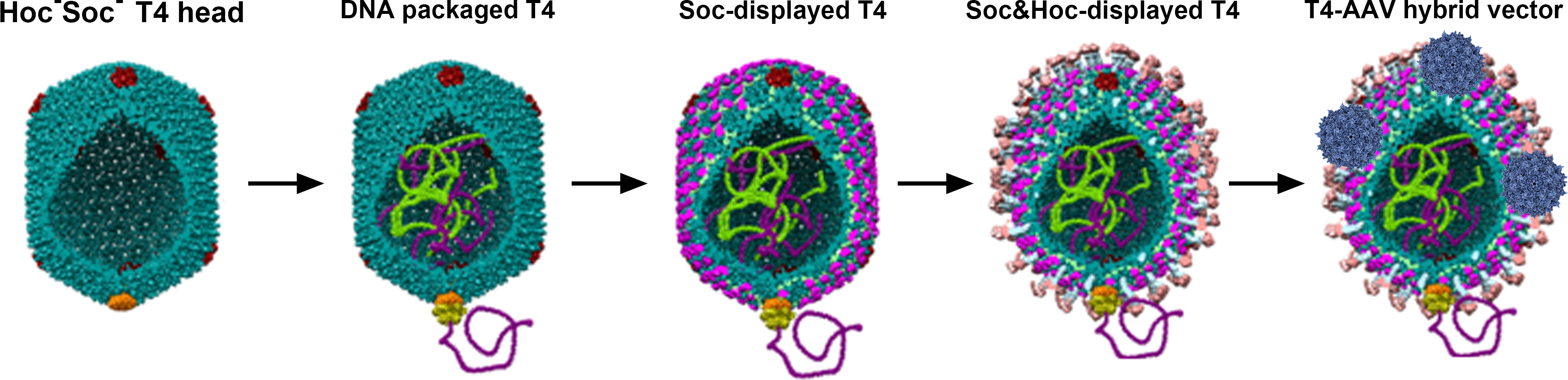
Figure 1. Schematic of DNA & protein carried T4 and T4-AAV hybrid vector
In this protocol, we describe the construction, expression, purification, and biotinylation of Soc and BAP-Hoc (BAP, Biotin Acceptor Peptide), and the assembly of Soc-biotin-avidin and Hoc-BAP-biotin-avidin. Soc and BAP-Hoc were biotinylated in vitro using biotin N-Hydroxysuccinimide (NHS) esters and BirA ligase, respectively. NHS-activated biotins react efficiently with primary amino groups (-NH2) of Soc protein (the side chain of lysine residues and the N-terminus) to form stable amide bonds. BirA biotin ligase site-specifically biotinylates a lysine side chain within a 15-amino acid BAP peptide fused to the N terminus of Hoc protein. We also provide a description of the production, purification, and modification of AAV and T4 head. Finally, we describe the methodology for generating the hybrid T4-AAV particle using biotinylated AAV, T4 head, and the “linker” molecules, Soc-biotin-avidin and Hoc-BAP-biotin-avidin (Figure 2). This protocol describes the assembly of hybrid T4-AAV vector in ~2 weeks. Although this study focused on T4 and AAV to conceptually demonstrate the utility of combining distinct viral building blocks, in a broader context, other hybrid vectors containing the T4 ‘carrier’ complexed with eukaryotic viruses, bacteria, or polymers using bioaffinity attachment technology can be envisioned for delivery of multivalent gene and protein therapeutics (Akin et al., 2007; Yoo et al., 2011).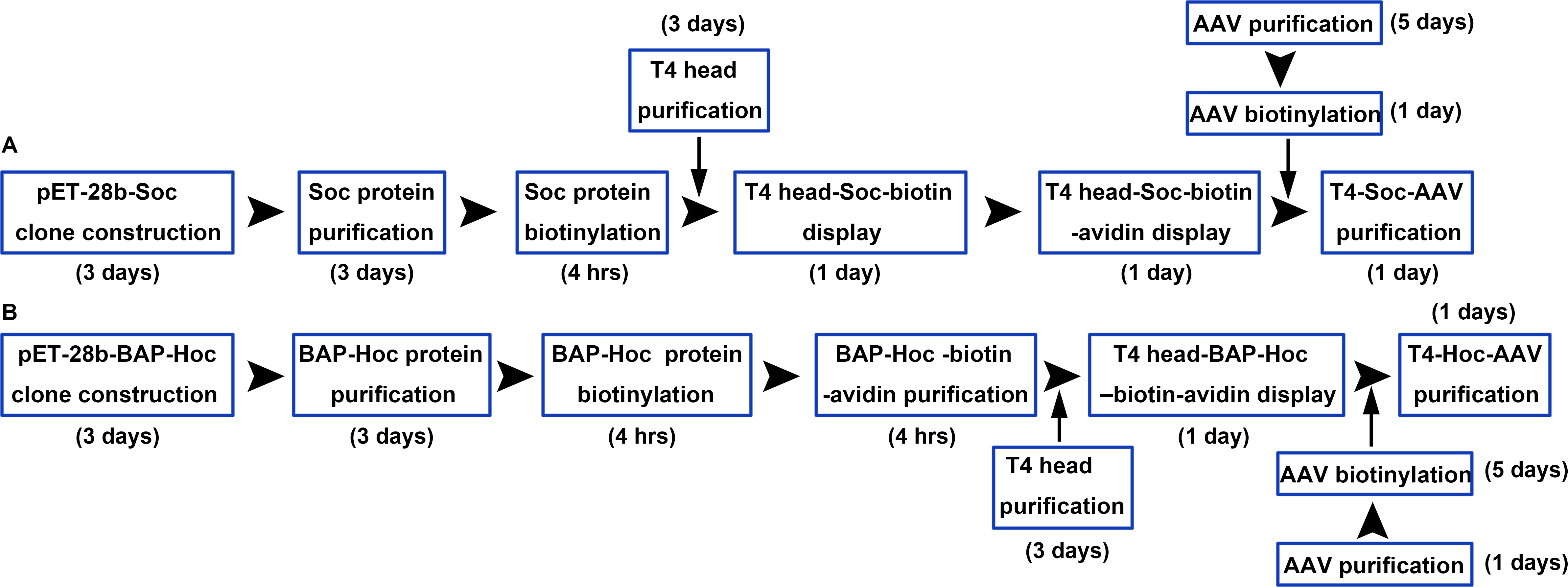
Figure 2. Workflow for construction of T4-Soc-AAV (A) and T4-Hoc-AAV (B) vectors. The T4 capsid lattice is decorated with two nonessential outer capsid proteins, Soc and Hoc. Soc and BAP-Hoc proteins were expressed in E.coli, purified by chromatography, and biotinylated. Two bridge molecules were constructed to attach AAV to the T4 head, Soc-biotin-avidin and Hoc-biotin acceptor peptide (BAP)-biotin-avidin. Soc and Hoc attach to the T4 head, and avidin attaches to biotinylated AAV (bioAAV) on the other side. T4-Soc-AAV (A) and T4-Hoc-AAV (B) conjugates were then purified by Ni2+-NTA agarose chromatography.
Materials and Reagents
- 15 ml tubes (GeneMate, catalog number: C3394-1 )
- 500 ml flask (VWR, catalog number: 89095-276 )
- 50 ml tubes (CELLSTAR, catalog number: 227270 )
- 125 ml flask (VWR, catalog number: 89095-258 )
- 15 ml Corex glass tubes (Corning)
- 1.7 ml sterile Eppendorf tube (Eppendorf, catalog number: 022431021 )
- Millex-GV Syringe Filter Unit, 0.22 µm (Millipore-Sigma, catalog number: SLGV033RS )
- HisTrap HP histidine-tagged protein purification columns (GE Healthcare, catalog number: 17524701 )
- Hi-load 16/600 Superdex200pg Gel filtration chromatography column (GE Healthcare, catalog number: 28989335 )
- Zeba Spin Desalting Columns (Thermo Scientific, catalog number: 89893 )
- Amicon Ultra-4 Centrifugal Filter Units (Millipore-Sigma, catalog number: UFC810008 )
- Competent E. coli DH5α cells (New England Biolabs, catalog number: C2987 )
- Competent E. coli BL21-CodonPlus (DE3)-RIPL cells (Agilent Technologies, catalog number: 230280 )
- HEK293T cells (ATCC, catalog number: ACS-4500 )
- Plasmids
a.pET-21a-BirA (Addgene, catalog number: 20857 )
b.pAAV-DJ (Cell Biolabs, catalog number: VPK-420-DJ )
c.pAAV-Helper (Cell Biolabs, catalog number: VPK-400-DJ )
d.pAAV-GFP (Cell Biolabs, catalog number: AAV-400 )
e.pAAV-luciferase (Cell Biolabs, catalog number: AAV-321 )
f.pAAV-mCherry (Cell Biolabs, catalog number: VPK-410 ) - Luria-Bertani (LB) medium (Quality Biological, catalog number: 10128-254 )
- Kanamycin (Gold Biotechnology, catalog number: K-120 )
- 2x Phusion High-Fidelity PCR Master Mix (Thermo Scientific, catalog number: F531S )
- Restriction enzymes
a.FastDigest NheI (Thermo Scientific, catalog number: FD0973 )
b.FastDigest HindIII (Thermo Scientific, catalog number: FD0504 )
c.FastDigest BglII (Thermo Scientific, catalog number: FD0084 )
d.FastDigest XhoI (Thermo Scientific, catalog number: FD0694 ) - FastAP Thermo sensitive Alkaline Phosphatase (Thermo Scientific, catalog number: EF0654 )
- T4 DNA Ligase (Thermo Scientific, catalog number: EL0014 )
- AccuGENETM 10x TBE Buffer (Lonza, catalog number: 50843 )
- QIAquick Gel Extraction Kit (QIAGEN, catalog number: 28706 )
- QIAprep Spin Miniprep Kit (QIAGEN, catalog number: 27106 )
- CompactPrep Plasmid Midi Kit (QIAGEN, catalog number: 12843 )
- Chloramphenicol (Amresco, catalog number: 0230-100G )
- Complete proteinase inhibitor cocktail (Roche, catalog number: 4693159001 )
- Isopropyl β-D-1-thiogalactopyranoside (IPTG) (Gold Biotechnology, catalog number: 12481C )
- EDTA (Quality Biological, catalog number: 351-027-721 )
- Dimethyl sulfoxide (DMSO) (ATCC, catalog number: 80713185 )
- Sodium acetate (Sigma-Aldrich, catalog number: 229873 )
- Ethanol (VWR, catalog number: BDH 1156-4LP )
- Deoxyribonuclease I (DNase I) (Sigma-Aldrich, catalog number: D5025 )
- Benzonase endonuclease (Millipore-Sigma, catalog number: 1016540001 )
- M9CA medium powder (Amresco, catalog number: J864-500G )
- Proteinase K Solution (Thermo Scientific, catalog number: AM2546 )
- HiTrap DEAE Sepharose FF (GE Healthcare, catalog number: 17515401 )
- HiTrap AVB Sepharose HP (GE Healthcare, catalog number: 28411211 )
- Bovine Serum Albumin, Acetylated (Promega, catalog number: R3961 )
- 4-20% Tris-Glycine Mini Gels (Thermo Scientific, catalog number: XP04205BOX )
- Coomassie Brilliant Blue R-250 Staining Solution (BIO-RAD, catalog number: 1610436 )
- Chloroform, HPLC Grade (Thermo Scientific, catalog number: AA43685M1 )
- Slide-A-Lyzer Dialysis Cassettes (Thermo Scientific, catalog number: 66380 )
- PBS (10x), pH 7.4 (Quality Biological, catalog number: 50146771 )
- Lipofectamine 2000 (Thermo Scientific, catalog number: 11668027 )
- QuickTiter AAV Quantitation Kit (Cell Biolabs, catalog number: VPK-145 )
- EZ-Link NHS-Biotin Reagents (Thermo Scientific, catalog number: 20217 )
- Dulbecco's Modified Eagle Medium (DMEM) (Thermo Scientific, catalog number: 10313039 )
- Sodium Chloride, 5 M (Quality Biological, catalog number: 351036491 )
- Ni-NTA Agarose (QIAGEN, catalog number: 30210 )
- Biotin (Sigma-Aldrich, catalog number: B4639 )
- ATP (Sigma-Aldrich, catalog number: A1852-1VL )
- pET-21a-BirA (Addgene, catalog number: 20857 )
- Tryptone (GROWCELLS, catalog number: MCMA-0901 )
- Yeast extract (Affymetrix, catalog number: 8013-01-2 )
- NaCl (Affymetrix, catalog number: 21618-5KG )
- Dextrose (AMRESCO, catalog number: 0188-IKG )
- Na2HPO4 (VWR, catalog number: 0404-2.5KG )
- KH2PO4 (Alfa Aesar, catalog number: 11594 )
- Tris-HCl (Quality Biological, catalog number: 351-006-101 )
- Imidazole (Alfa Aesar, catalog number: 11594 )
- Methanol (VWR, catalog number: BDH 1135-4LP )
- Acetic acid (VWR, catalog number: BDH3908-3.8LP )
- MgSO4 (Sigma-Aldrich, catalog number: M7506 )
- Moore’s medium (see Recipes)
- 1 M IPTG (see Recipes)
- HisTrap binding buffer (see Recipes)
- HisTrap elution buffer (see Recipes)
- Gel filtration buffer (see Recipes)
- SDS-PAGE destaining solution (see Recipes)
- M9CA medium (see Recipes)
- PI-Mg buffer (see Recipes)
- Head binding buffer (see Recipes)
- Head elution buffer (see Recipes)
- DNA packaging buffer (see Recipes)
- PBS-MK buffer (see Recipes)
- DMEM cell culture complete medium (see Recipes)
Equipment
- Pipettes (VWR, catalog number: 89130898 )
- Water bath (VWR, model: 1217 )
- -80 °C freezer (Panasonic, model: MDF-U76VC )
- SS34 rotor (SORVALL)
- GS3 rotor (SORVALL)
- AM 2.18 rotor
- PCR machine (BIO-RAD, model: T100 thermal cycler )
- Sorvall RC-5C plus centrifuge (Thermo Scientific, model: SorvallTM RC-5C plus )
- French press (Thermo Scientific)
- AKTA-prime system (Amersham Biosciences)
- AKTA Fast Protein Liquid Chromatography (FPLC) system (GE Healthcare, model: UPC-900 )
- NanoDrop 2000 spectrophotometer (Thermo Scientific, model: NanoDropTM 2000 )
- Shaking incubator (New Brunswick Scientific, model: Innova 43 )
- ChemiDoc Imaging Systems (BIO-RAD, 731BR02947 )
- MilliQ water machine (Millipore-Sigma, lot number: F8BA18025 )
- Bench centrifuge (Eppendorf, model: 5415D )
- Autoclave (Tuttnauer, model: 3870EA-B/L )
- High-speed multi-function centrifuge (Jouan, model: MR-23i )
Software
- ImageJ (National Institutes of Health Image, https://imagej.nih.gov/ij/)
- Quantity One software (Bio-Rad, CA)
- SnapGene (GSL Biotech LLC)
Procedure
- Plasmid construction (3 days)
Hoc and/or Soc can be efficiently displayed on the T4 capsid as the linker for AAV attachment. Here, we constructed pET-28b-Soc and pET-28b-BAP-Hoc for Hoc/Soc protein expression. Additionally, the BirA-expressing plasmid pET-21a-BirA was purchased from Addgene (plasmid no. 20857 ). Plasmids pAAV-DJ (# VPK-420-DJ ), pAAV-Helper (# VPK-400-DJ ), pAAV-GFP (# AAV-400 ), pAAV-luciferase (# AAV-321 ), and pAAV-mCherry (# VPK-410 ) were purchased from Cell Biolabs.- pET-28b-Soc construction (3 days)
- Amplify RB69 Soc gene fragment (228 bp) from RB69 phage genomic DNA using Thermo Fisher 2x Phusion high-fidelity PCR master mix according to the manufacturer’s protocol (https://www.thermofisher.com/order/catalog/product/F531S#/F531S).The primers are shown as follows, where the underlined sequences indicate the recognition sequences for the respective restriction enzymes:
Soc NheI Forward:
5′-GCATCCGCTAGCGGTGGTTATGTA A ACATCAAA-3′.
Soc XhoI Reverse:
5′-GTGCTCGAGCACCACTTACTGGTGTAGGGGTAAAC-3′. - Add 1/10 volume of 10x DNA loading buffer to each PCR sample and load 50 μl reaction onto the 1.5% agarose-TBE gel, and separate them by agarose gel electrophoresis. Additionally, load the 100 bp plus DNA marker for size verification.
- Cut the DNA band of expected size using a clean blade, purify Soc DNA using QIAGENQIA quick gel extraction kit according to the manufacturer’s protocol.
- Digest 1 μg purified Soc PCR product with NheI and XhoI by following the manufacturer’s instructions with incubation at 37 °C for 4 h. Meantime, digest 1 μg pET-28b vector plasmid with NheI and XhoI at 37 °C for 4 h.
- To prevent vector recircularization during ligation, dephosphorylate the 5′-ends of the digested pET-28b with 1 μl Fast AP thermos sensitive alkaline phosphatase without changing the restriction enzyme buffer and incubate at 37 °C for 40 min.
- Separate the digested Soc insert and pET-28b vector using 1.5% and 1% agarose gel electrophoresis, respectively. Then purify them using the QIAGENQIA quick gel extraction kit according to the manufacturer’s instructions. Store the digested insert and vector at -20 °C until needed.
- Ligate insert and vector at a molar ratio of 5:1 using T4 DNA ligase at 22 °C for 2 h (in a water bath). Prepare the vector backbone without the insert as a negative control to evaluate vector recircularization.
- Transform E. coli DH5α competent cell with the ligation product using heat shock according to the manufacturer’s instructions and incubate on the kanamycin-LB plate overnight at 37 °C.
- Pick 2-3 single colonies and inoculate each into 3 ml LB medium supplemented with 50 μg/ml of kanamycin in 15 ml tubes. Incubate the tubes in a shaking incubator overnight at 220 rpm and 37 °C. Purify plasmid DNA from the above cultures using the QIAGEN plasmid mini kit as per the manufacturer’s protocol.
- Sequence clones with T7 and T7ter sequencing primers to confirm the proper insertion of Soc sequence into the pET-28b backbone. The resulting pET-28b-Soc plasmid should contain hexa-histidine tag at the both NH2- and COOH- termini of Soc.
- Amplify RB69 Soc gene fragment (228 bp) from RB69 phage genomic DNA using Thermo Fisher 2x Phusion high-fidelity PCR master mix according to the manufacturer’s protocol (https://www.thermofisher.com/order/catalog/product/F531S#/F531S).The primers are shown as follows, where the underlined sequences indicate the recognition sequences for the respective restriction enzymes:
- pET-28b-BAP-Hoc construction (3 days)
- Perform two rounds of PCR reactions to amplify BAP-Hoc from T4 genomic DNA using 2x Phusion high-fidelity PCR master mix and the following three primers. The underlined sequences indicate the recognition sequences for the respective restriction enzymes.
BAP-Hoc Forward 1:
5′-ATCGAGTGGCACGAGGGTCTTTCGATGACTTTTACAGTTGATATAACTCC-3′
BAP-Hoc NheI Forward 2:
5′-TTCTAGCTAGCGGTCTTAACGACATCTTCGAGGCACAGAAGATCGAGTGGCACGAGGGTCTTTCG-3′
BAP-Hoc HindIII Reverse:
5′-ATAAAGCTTTTATGGATAGGTATAGATGATACCAGTTTC-3′
To label Hoc protein with biotin, we genetically fused biotin acceptor peptide (BAP) to the N-terminus of Hoc. Perform the first round of PCR by fusing Hoc to part of the BAP sequence using the BAP-Hoc Forward 1 and BAP-Hoc HindIII Reverse primers. Obtain full-length BAP-Hoc (1,197 bp) by the second round of PCR using the BAP-Hoc NheI Forward 2 and BAP-Hoc HindIII Reverse primers. - Digest BAP-Hoc PCR product and pET-28b vector with Nhe I and Hind III. Then, sub-clone the digested BAP-Hoc fragment into the pET-28b vector according to the Steps A1b to A1j. The resulting pET-28b-BAP-Hoc plasmid should contain Hoc in-frame fusion with BAP, a 15-aa vector biotin acceptor peptide (GLNDIFEAQKIEWHE) at the NH2-terminus of Hoc, and also a hexa-histidine sequence at the COOH-terminus of Hoc.
- Perform two rounds of PCR reactions to amplify BAP-Hoc from T4 genomic DNA using 2x Phusion high-fidelity PCR master mix and the following three primers. The underlined sequences indicate the recognition sequences for the respective restriction enzymes.
- pET-28b-Soc construction (3 days)
- Protein expression, purification, and modification (4 days)
- Soc and BAP-Hoc protein expression and purification (3 days)
- Transform 15 ng of pET-28b-Soc or pET-28b-BAP-Hoc into 20 μl BL21-CodonPlus (DE3)-RIPL competent cells using the heat-shock method according to the manufacturer’s protocol. BL21-CodonPlus(DE3)-RIPL cells contain a ColE1-compatible, pACYC-based plasmid that has chloramphenicol resistance for protein expression. Plate the transformed cells on LB-kanamycin/chloramphenicol plates (50 μg/ml kanamycin and 30 μg/ml chloramphenicol) and incubate overnight at 37 °C.
- Pick a single colony and inoculate into a 500 ml flask containing 50 ml Moore’s medium containing 50 μg/ml kanamycin and 30 μg/ml chloramphenicol. Incubate overnight in a shaking incubator at 220 rpm and 37 °C.
- Inoculate 20 ml of overnight cultures into 1 L of Moore’s medium containing 50 μg/ml kanamycin and 50 μg/ml chloramphenicol. Incubate the cultures at 220 rpm and 37 °C until the cell density reaches OD600 = 0.4-0.5. Then, cool down the temperature of the cultures to 28 °C in a shaking incubator for 40 min at 220 rpm before adding IPTG.
- Add 1 ml 1 M IPTG to the 1 L cultures (final concentration 1 mM) and incubate for 2-3 h at 28 °C and 220 rpm for inducing protein expression.
- Harvest the induced cells by centrifuging the cultures using 500 ml centrifuge bottles and GS3 rotor of Sorvall RC-5C plus centrifuge at 7,000 rpm (8,288 x g) for 10 min at 4 °C. Discard the supernatant and store the pellet at -80 °C until purification.
- Resuspend the pellet with 50 ml HisTrap binding buffer containing one pill of complete proteinase inhibitor cocktail on ice. Lyse the cells by French press at 10,000 psi twice.
- Centrifuge the cell lysate using 30 ml tubes and SS34 rotor in Sorvall RC-5C plus centrifuge at 18,000 rpm (38,724 x g) for 30 min at 4 °C.
- Discard the pellet and harvest the supernatant containing soluble Soc or BAP-Hoc protein. Filter the supernatant through 0.22 μm Millipore filter units before loading onto the HisTrap column.
- Set up 1 ml HisTrap HP column on AKTA-prime system. Wash the column with 30 ml of water, followed by 30 ml of HisTrap binding buffer to equilibrate the column.
- Load the filtered supernatant onto the HisTrapHP column at a flowing speed of 1 ml/min (Increased flow rate will decrease protein binding to column.), followed by washing the loaded column with 50 ml HisTrap binding buffer.
- Set up the elution procedures of the HisTrap HP column on the AKTA-prime system. The parameters are as follows: Concentration (% Buffer B): 0; Gradient Length: 40; Target (% Buffer B): 100; Flow Rate: 1 ml/min; Fraction Base: ml; Fraction Size: 1; Pressure Limit: 0.3. Using the HisTrap binding buffer for pump A and the HisTrap elution buffer for pump B, elute the His-tag protein with a 20-400 mM linear imidazole gradient.
- Collect and pool the peak fractions, and concentrate the pool to 4-5 ml using Amicon Ultra-4 centrifugal filtration (3 kDa cutoff for Soc and10 kDa cutoff for BAP-Hoc) according to the manufacturer’s protocol.
- Set up AKTA Fast Protein Liquid Chromatography (FPLC) system to further purify by size-exclusion chromatography as well as desalt Soc or BAP-Hoc proteins. Wash the Hi-load 16/600 Superdex 200 column with 150 ml gel filtration buffer. Load the concentrated His-tag protein (4-5 ml) onto the Hi-load 16/600 Superdex 200 column with a flow rate of 1.0 ml/min and up to 0.5 MPa pressure.
- Collect and pool the peak fractions from gel filtration elution based on the molecular size, and concentrate using Amicon Ultra-4 centrifugal filtration.
- Check purity by SDS-PAGE and quantify the concentration of the Soc and BAP-Hoc protein using NanoDrop. The concentration measurement parameters are as follows: Soc, molecular size 10.5 KDa, Extinction coefficients 11,460, Abs 0.1% (= 1 g/L) 1.088; BAP-Hoc, molecular size 44.9 KDa, Extinction coefficients 70945, Abs 0.1% (= 1 g/L) 1.580. Aliquot the concentrated protein and store at -80 °C until needed.
- Soc protein biotinylation (4 h)
Biotin N-Hydroxysuccinimide (NHS) esters are the most popular type of biotinylation reagent. NHS-activated biotins react efficiently with primary amino groups (-NH2) of protein (the side chain of lysine residues and the N-terminus of each polypeptide) to form stable amide bonds.- Exchange Soc protein gel filtration buffer (Tris buffer) with 1x PBS buffer (pH 7.4) using 10 ml Zeba Spin Desalting Columns (7K MWCO) according to the manufacturer’s instructions. Avoid buffers containing primary amines (e.g., Tris or glycine) as these will compete with the biotinylation reaction. Proteins in Tris or other amine-containing buffers must be exchanged into an amine-free buffer at pH 7.2-8.0. Concentrate Soc protein in PBS buffer to ~5 mg/ml using Amicon Ultra-4 centrifugal filtration.
- Remove the vial of NHS-Biotin reagents from -20 °C storage and fully equilibrate it to room temperature for 30 min before opening to avoid moisture condensation inside the container. NHS-Biotin reagents are moisture-sensitive.
- Immediately before use, dissolve 2.0 mg NHS-Biotin reagent in 590 μl of dimethyl sulfoxide (DMSO) solvent to prepare a 10 mM solution.
- Add 500 μl of 10 mM biotin reagent solution to the 500 μl 5 mg/ml Soc protein solution (20-fold molar excess). Incubate reaction on ice for 2 h.
- Purify the labeled Soc protein using 10 ml Zeba Spin Desalting Columns (7K MWCO) according to the manufacturer’s protocol. Finally, elute the Soc-biotin protein in 1x PBS buffer (pH 7.4). Aliquot the labeled protein and store at -80 °C until needed.
- Hoc-BAP-Biotin-Avidin (HBBA) production (8 h)
- Exchange Hoc-BAP protein gel filtration buffer with 1x PBS-MgCl2 buffer (pH 7.4) using 10 ml Zeba Spin Desalting Columns (7K MWCO) according to the manufacturer’s instructions.
- Add 1 μM recombinant BirA ligase to 30 μM Hoc-BAP monomer in the presence of 0.3 mM biotin and 5 mM ATP for Hoc-BAP biotinylation. Let the reaction proceed at room temperature for 3 h with gentle mixing on a rocking platform for Hoc-BAP-biotin formation (HBB).
- Remove the excess biotin using 10 ml Zeba Spin Desalting Columns (7K MWCO). Confirm biotinylation by streptavidin-HRP Western blot analysis.
- Incubate Hoc-BAP-biotin with avidin at a 1:3 ratio for 1 h at 4 °C to form Hoc-BAP-biotin-avidin complex.
- Set up AKTA-FPLC system. Wash the Hi-load 16/600 Superdex 200 column with 150 ml gel filtration buffer. Load the biotinylation reaction mixture containing HBB, HBBA, avidin, BirA, and Hoc-BAP onto the Hi-load 16/600 Superdex 200 column with a flow rate of 1.0 ml/min.
- Collect and pool the peak fractions of HBBA from gel filtration elution based on the molecular size, and concentrate using Amicon Ultra-4 centrifugal filtration. Check the purity by SDS-PAGE. Aliquot the concentrated protein and store at -80 °C until needed.
- Soc and BAP-Hoc protein expression and purification (3 days)
- Bacteriophage T4 head production (3 days)
- Streak out the glycerol stock of E. coli P301 cells on the LB-agar plate and leave it at 37 °C overnight. Pick a single colony and inoculate into 20 ml sterile LB + M9A medium (equal volumes of LB and M9A) in a 125 ml flask. Grow at 37 °C for 6-8 h.
- Inoculate 5 ml fresh E. coli P301 culture into LB + M9A medium (500 ml) at a dilution of 1:100. Leave the cap loose to allow aeration and grow the culture in the shaking incubator at 37 °C to a density of 4 x 108 cells/ml (in the cell counter, if each small square has 5 cells/ml, the cell density is equivalent to 108 cells/ml).
- Infect the culture with 10-amber.13-amber.hoc-del.soc-delmutant T4 at an M.O.I. (number of phage added/each bacterial cell) of 4. It should be noted that phage T4 has no tropism to human cells, has no known toxicity or pathogenicity, and exhibits no preexisting immunity. The safety of T4 phage application in humans was reported. This involves simply adding a calculated volume of the phage solution into the bacterial culture. Mix the culture as you are adding the phage solution so that the phage is uniformly distributed into the culture. Put the culture back in the shaking incubator for 5 min.
- Superinfect at the same M.O.I. as above (a second T4 infection). Put the culture back in the shaking incubator and grow for 32 min more. At the 30th min quickly check the cell number under the microscope to see if the cell number begins to reduce. If the cell number is less than 4 x 108 cells/ml, just stop the infection and do centrifugation. If the cell number has not changed, leave the culture at 37 °C for 1-2 min more (Don’t leave for more than 35 min. The cells would have lysed). Reserve the Sorvall centrifuge ahead of time so that it is available when you need it.
- Distribute equally into two 500 ml centrifuge bottles. Centrifuge the cells at 7,000 rpm (8,288 x g) for 12 min at 4 °C in the Sorvall RC-5C using GS3 rotor. Discard the supernatant.
- To the pellet, add 50 ml (25 ml to each bottle) of PI-Mg buffer plus 20 μg/ml DNAaseI. Resuspend the viscous pellet by repeated pipetting. Add 1 ml chloroform and incubate at 37 °C for 30 min in a shaker at 220 rpm to lysis cells and digest DNA.
- Collect the supernatant by centrifuging at 4,300 x g or 12 min. Discard the pellet. Transfer the supernatant to four 15 ml Corex glass tubes (Be careful to remove the supernatant using disposable pipettes). Centrifuge the supernatant using a SS34 rotor in Sorvall RC-5C plus centrifuge at 18,000 rpm (38,724 x g) for 45 min at 4 °C (using the rubber adapters). Discard the supernatant. The pellet mainly contains T4 viral heads.
- Add 500 μl of PI-Mg buffer to each pellet. Leave the pellet overnight at 4 °C. Resuspend the pellet. Break the pellet with the tip of the pipette to gently resuspend it. Remove the four tubes pellet into one tube.
- Conduct second-round low- and high-speed centrifugations. Resuspend the pellet with 1 ml PI-Mg buffer. Then add 50 ml of head binding buffer (no NaCl) for DEAE column purification.
- Filter the sample using a 0.22 μm filter unit. Load the sample onto a 5 ml DEAE column (1 ml/min) pre-equilibrated with 100-150 ml of head binding buffer (no NaCl). Elute with a linear gradient from 0 mM to 300 mM NaCl (40 ml) in the head binding buffer. Collect the peak fractions (1 ml each).
- Concentrate the purified heads using Amicon Ultra-4 centrifugal filtration.
- Check purity and concentration by SDS-PAGE. Mix 1/5 volume of T4 head or BSA marker with 5x SDS loading buffer and boil for 10 min. Load 1, 2, 3, and 4 μl of T4 head, as well as 1, 2, 4, and 8 μg of BSA on a 4-20% Tris-Gly SDS-PAGE gel, and electrophorese according to the manufacturer’s instructions.
- Stain the gel with Coomassie blue R-250 staining solution, microwave for 1 min, and keep shaking at room temperature for 30 min.
- Remove the staining solution, add destain solution, microwave for 1 min, and keep shaking at room temperature until the background becomes clean.
- Scan the protein gel with ChemiDoc Imaging Systems and quantify the protein bands with Quantity One software according to the manufacturer’s instructions. Calculate the concentration of T4 head based on the BSA standard curve generated by Microsoft Excel.
- Aliquot the T4 heads and store at -80 °C until needed.
- In vitro DNA packaging and protein display of the T4 head (2 days)
- T4 DNA packaging (1 day)
- Prepare plasmid DNA pAAV-GFP or pAAV-Luciferase from the corresponding E. coli cultures using the QIAGEN plasmid midi kit as per the manufacturer’s protocol.
- Linearize pAAV-GFP or pAAV-Luciferase using restriction enzyme XbaI or BglII for T4 head packaging according to the manufacturer’s protocol.
- Purify the linearized plasmid DNA.
- Add one volume of 8 M sodium acetate (-20 °C) to the enzyme cutting reaction.
- Add two-volume of room temperature isopropanol and keep on ice for 30 min.
- Centrifuge at 18,000 rpm (38,724 x g) for 30 min at 4 °C.
- Discard the supernatant and rinse with 70% Ethanol.
- Centrifuge at 18,000 rpm (38,724 x g) for 30 min at 4 °C.
- Discard the supernatant and air dry pellet (20 min) and dissolve the pellet in DNA elution buffer.
- Quantify the concentration of linearized plasmid DNA using NanoDrop.
- Perform in vitro DNA packaging. Each 20 μl of reaction mixture contains 3 μM full-lengthgp17, linearized plasmid DNA (DNA:heads = 20:1, 2.4 μg pAAV-GFP or 2.8 μg pAAV-Luciferase), 2 μl 10x packaging buffer, and autoclaved MilliQ water. Incubate the mixture at 37 °C for 5 min. Then add 2 x 1010 T4 head and incubate at 37 °C for 30 min.
- Add 1 μl benzonase nuclease to the packaging reaction and incubate at 37 °C for 20 min to remove excess unpackaged DNA.
- Check the T4 head packaging efficiency. Release the encapsidated nuclease-resistant DNA by treatment with 50 mM EDTA, 0.5 μg/μl proteinase K, and 0.2% SDS for 30 min at 65 °C. Analyze the packaged DNA by 1% agarose gel electrophoresis.
- Quantify the amount of packaged DNA using Quantity One software. Define the packaging efficiency as the number of DNA molecules packaged per T4.
- Keep DNA-packaged T4 head on ice for further protein display assay.
- T4 protein display (1 day)
- Add Soc-biotin or HBBA protein (20:1 ratio of Soc-biotin to T4 870 Soc-binding-sites; 20:1 ratio of HBBA to T4 155 Hoc-binding-sites) (Fokine et al., 2004) to the DNA-encapsidated T4 heads as described above, gently vortex to mix, and incubate at 4 °C for 1 h.
- Sediment the T4 head particles with Soc-biotin or HBBA protein bound at 18,000 rpm (32,150 x g) for 1 h at 4 °C using AM 2.18 rotor in Jouan MR-23i centrifuge or equivalent.
- Discard the supernatant to remove the unbound protein.
- Wash the T4 head pellet containing the bound Soc-biotin or HBBA protein twice with 1x PBS buffer.
- Incubate the pellets at 4 °C overnight and then resuspend in 1x PBS.
- Analyze protein-displayed T4 heads by SDS-PAGE as described in procedures from Steps C12 to C15. Determine the density volumes of Soc-biotin or HBBA and gp23* (major capsid protein) bands for each lane, and calculate the copy number of Soc-biotin or HBBA molecules per capsid using the known copy number of gp23*, which is 930 copies per head.
- T4 DNA packaging (1 day)
- Recombinant adeno-associated virus (AAV) production and biotinylation (6 days)
- AAV production (5 days)
- One day before transfection, plate sufficient HEK293T cells (~3 x 106, 12 ml DMEM complete medium) to 100 mm cell culture dishes to achieve 70-80% confluence on the day of transfection.
- Co-transfect cells with pAAV rep/cap expression vector, pAAV-luciferase, and pAAV-Helper with the ratio of vectors at 1:1:1 (15 μg pAAV-DJ: 15 μg pAAV-Luciferase: 15 μg pAAV-Helper) using Lipofectamine 2000 according to the manufacturer’s protocol.
- Seventy-two hours after transfection, add 240 μl 0.5 M EDTA (to a final of 10 mM) to the 12 ml DMEM cultures and incubate for 5 min at room temperature. Gently shake the culture dish several times and harvest all media, including cells, in a sterile 50 ml tube.
- Centrifuge the cell suspension at 150 x g for 5 min. Remove the supernatant and resuspend the cell pellet in 0.5 ml sterile 1x PBS.
- Freeze and thaw the cell suspension four times by placing it alternately in a dry ice/ethanol bath and a water bath of 37 °C. Remove cell debris by centrifugation at 10,000 x g for 10 min and collect the supernatant as AAV crude lysate.
- Purify AAV by HiTrap AVB Sepharose HP column according to the manufacturer’s instructions (GE Healthcare, IL). Collect and pool peak fractions.
- Dialyze AAV pool against PBS-MK buffer overnight in Slide-A-Lyzer dialysis cassettes as per the manufacturer’s protocol. Concentrate AAV particles using Amicon Ultra-4 centrifugal filtration (100 KDa size cutoff). Determine AAV titers using a QuickTiter AAV Quantitation kit (Cell Biolabs, CA) according to the manufacturer’s instructions. Aliquot and store the purified AAV at -80 °C.
- AAV biotinylation (1 day)
- Remove the vial of NHS-Biotin reagents from -20 °C storage and fully equilibrate it to room temperature for 30 min before opening to avoid moisture condensation inside the container.
- Immediately before use, dissolve 4.0 mg NHS-Biotin reagent in 590 μl of DMSO solvent to prepare a 20 mM solution.
- Add 590 μl of 20 mM biotin reagent solution to the 500 μl AAV solution (~1012 particles). Incubate reaction on ice for 2 h.
- Add 100 mM glycine to quench biotin reagent.
- Dialyze labeled AAV against PBS-MK buffer overnight in Slide-A-Lyzer dialysis cassettes as per the manufacturer’s protocol to remove excess biotin reagent and byproducts. Confirm biotinylation by streptavidin-HRP Western blot analysis. Aliquot the labeled AAV and store at -80 °C until needed.
- AAV production (5 days)
- T4-AAV hybrid vector production (1 day)
- Distribute 1 ml Ni2+-NTA (nitrilotriacetic acid) agarose beads into a 1.7 ml sterile Eppendorf tube. Centrifuge the beads at 100 x g for 1 min. Remove the supernatant and wash the beads pellet with 1 ml HisTrap binding buffer three times. Resuspend the beads pellet in 0.1 ml HisTrap binding buffer after washing.
- Add 0.2 ml 1012 T4-Soc-biotin or T4-HBBA particles into Ni2+-NTA agarose beads and incubate for 1 h with gentle shaking at 4 °C.
- Centrifuge the mixture at 100 x g for 30 s. Remove the supernatant and gently wash the T4-Soc-biotin or T4-HBBA bound beads with 1 ml HisTrap binding buffer five times. Resuspend the beads pellet in 0.1 ml HisTrap binding buffer after washing.
- For assembly of T4-Soc-AAV assembly, dissolve avidin in HisTrap binding buffer and then add to the T4-Soc-biotin bound beads (1:1 of avidin to T4 870 soc-binding-sites). Incubate for 20 min with gentle shaking at 4 °C. Centrifuge the mixture at 100 x g for 30 s. Remove the supernatant and gently wash the T4-Soc-biotin-avidin bound beads with 1 ml HisTrap binding buffer five times. Resuspend the beads in 0.1 ml HisTrap binding buffer after washing.
- Add the biotin-labeled AAV vectors (sub-heading E-2) (4:1 of AAV to T4 particles) to the T4-Soc-biotin-avidin or T4-HBBA immobilized beads and incubate for 30 min with gentle shaking at 4 °C.
- Remove the supernatant and gently wash the T4-Soc-AAV or T4-Hoc-AAV bound beads with 1 ml HisTrap binding buffer five times. Resuspend the beads pellet in 0.1 ml HisTrap elution buffer after washing. Incubate for 5 min with gentle shaking at 4 °C.
- Dialyze T4-AAV hybrid viral complex vector against PBS-MK buffer overnight in Slide-A-Lyzer dialysis cassettes as per the manufacturer’s protocol. Aliquot and store at -80 °C until needed.
Data analysis
- Soc has five lysines, which provide primary amine groups. Hence, the purified Soc protein was efficiently biotinylated by activation with NHS-biotin reagent, which was confirmed by Western blot using streptavidin–horseradish peroxidase (HRP) against biotin (see Figures 2A and 2B in our original Science Advances research article) (Zhu et al., 2019).
- A biotin acceptor peptide BAP (GLNDIFEAQKIEWHE) was genetically fused to the N-terminus of T4 Hoc. Biotin ligase (BirA), which specifically recognizes a single lysine residue (L) in BAP, efficiently biotinylated BAP-Hoc protein in the presence of ATP and biotin (Schatz, 1993) (see Figures 3C and S1A in our original Science Advances research article). The ability of the modified BAP-Hoc to bind avidin indicated the biotinylation (see Figure S1B in our original Science Advances research article). Furthermore, the desired Hoc-avidin complex was finally purified by size-exclusion chromatography due to the significant molecular size difference between Hoc-avidin complex and other byproducts (see Figures S1C and S1D in our original Science Advances research article) (Zhu et al., 2019).
- Recombinant AAV-DJ vector was the chimera of a single type 2/type 8/type 9 and outperformed eight standard AAV serotypes in culture (Grimm et al., 2008). Triple-plasmid transfection and HiTrap AVB Sepharose HP column or iodixanol gradient ultracentrifugation were applied to produce and purify recombinant AAV-DJ vectors. The VP1, VP2, and VP3 subunits assemble into a complete 60-mer AAV capsid at a ratio of 1:1:10. VP subunits were labeled with biotin by the biotin-NHS reagent. Western blot using HRP-streptavidin indicated the presence of biotin conjugated to VP subunits in biotin-AAV-DJ vectors, while VP subunit bands were not detected in the unlabeled vectors (see Figures S3A and S3B in our original Science Advances research article) (Zhu et al., 2019).
- T4-Soc-Soc-Avidin or T4-HBBA particles were immobilized on a column of nickel beads through the hexa-histidine tag present at the terminus of Soc. This column allowed efficient capture of biotinylated AAV by the T4 Soc-avidin or HBBA bridge molecules and simultaneous removal of unbound AAV in the flow-through. The hybrid T4-AAV nanoparticles were finally eluted with imidazole and dialyzed against the PBS-MK buffer. The formation of T4-AAV nanoparticle was confirmed by western blot, the AAV amount in flow-through, and transmission electron microscopy (TEM) (see Figures 3 and S3 in our original Science Advances research article [Zhu et al., 2019]).
Recipes
- Moore’s medium
20 g tryptone
15 gyeast extract
8 g NaCl
2 g dextrose
2 g Na2HPO4
1 g KH2PO4
Add MilliQ H2O to 1 L - 1 M IPTG
238 mg IPTG- Add MilliQ H2O to 1 ml
- Aliquot and store at -20 °C
- HisTrap binding buffer
50 mM Tris-HCl pH 8.0
300 mM NaCl
20 mM imidazole
Store at 4 °C - HisTrap elution buffer
50 mM Tris-HCl pH 8.0
300 mM NaCl
400 mM imidazole
Store at at 4 °C - Gel filtration buffer
20 mM Tris-HCl pH 8.0
100 mM NaCl
Store at 4 °C - SDS-PAGE destaining solution
400 ml methanol
100 ml acetic acid
800 ml MilliQ water - M9CA medium
12.5 g M9CA medium powder
Add MilliQ H2O to 1 L - PI-Mg buffer
22 mM KH2PO4
70 mM NaCl
1 mM MgSO4
26 mM Na2HPO4 - Head binding buffer
50 mM Tris-HCl pH 8.0
5 mM MgSO4
Store at 4 °C - Head elution buffer
50 mM Tris-HCl pH 8.0
5 mM MgSO4
300 mM NaCl
Store at 4 °C - DNA packaging buffer
300 mM Tris-HCl pH 7.5
30 mM MgSO4
1 M NaCl
10 mM ATP
Store at -20 °C - PBS-MK buffer
1x PBS (pH 7.4)
1 mM MgCl2
2.5 mM KCl
Store at 4 °C - DMEM cell culture complete medium
435 ml DMEM
50 ml FBS
10 ml 1 M HEPES
5 ml 100x penicillin/streptomycin
Store at 4 °C
Acknowledgments
This work was supported by NIAID/NIH grant AI111538, and in part by NIH grant AI081726 and NSF grant MCB-0923873 to V.B.R. This protocol was adapted from our previously-published work (Zhu et al., 2019).
Competing interests
The authors declare no competing financial interests.
References
- Akin, D., Sturgis, J., Ragheb, K., Sherman, D., Burkholder, K., Robinson, J. P., Bhunia, A. K., Mohammed, S. and Bashir, R. (2007). Bacteria-mediated delivery of nanoparticles and cargo into cells. Nat Nanotechnol 2(7): 441-449.
- Fokine, A., Chipman, P. R., Leiman, P. G., Mesyanzhinov, V. V., Rao, V. B. and Rossmann, M. G. (2004). Molecular architecture of the prolate head of bacteriophage T4. Proc Natl Acad Sci U S A 101(16): 6003-6008.
- Fuller, D. N., Raymer, D. M., Kottadiel, V. I., Rao, V. B. and Smith, D. E. (2007). Single phage T4 DNA packaging motors exhibit large force generation, high velocity, and dynamic variability. Proc Natl Acad Sci U S A 104(43): 16868-16873.
- Ghosh, D., Lee, Y., Thomas, S., Kohli, A. G., Yun, D. S., Belcher, A. M. and Kelly, K. A. (2012). M13-templated magnetic nanoparticles for targeted in vivo imaging of prostate cancer. Nat Nanotechnol 7(10): 677-682.
- Grimm, D., Lee, J. S., Wang, L., Desai, T., Akache, B., Storm, T. A. and Kay, M. A. (2008). In vitro and in vivo gene therapy vector evolution via multispecies interbreeding and retargeting of adeno-associated viruses. J Virol 82(12): 5887-5911.
- Jain, A. and Cheng, K. (2017). The principles and applications of avidin-based nanoparticles in drug delivery and diagnosis. J Control Release 245: 27-40.
- Leffers, G. and Rao, V. B. (1996). A discontinuous headful packaging model for packaging less than headful length DNA molecules by bacteriophage T4. J Mol Biol 258(5): 839-850.
- Li, Q., Shivachandra, S. B., Zhang, Z. and Rao, V. B. (2007). Assembly of the small outer capsid protein, Soc, on bacteriophage T4: a novel system for high density display of multiple large anthrax toxins and foreign proteins on phage capsid. J Mol Biol 370(5): 1006-1019.
- Muzyczka, R. J. S. a. N. (2014). AAV-mediated gene therapy for research and therapeutic purposes. Annu Rev Virol (1): 427-451.
- Schatz, P. J. (1993). Use of peptide libraries to map the substrate specificity of a peptide-modifying enzyme: a 13 residue consensus peptide specifies biotinylation in Escherichia coli. Biotechnology (N Y) 11(10): 1138-1143.
- Shivachandra, S. B., Li, Q., Peachman, K. K., Matyas, G. R., Leppla, S. H., Alving, C. R., Rao, M. and Rao, V. B. (2007). Multicomponent anthrax toxin display and delivery using bacteriophage T4. Vaccine 25(7): 1225-1235.
- Stewart, M. P., Sharei, A., Ding, X., Sahay, G., Langer, R. and Jensen, K. F. (2016). In vitro and ex vivo strategies for intracellular delivery. Nature 538(7624): 183-192.
- Tao, P., Mahalingam, M., Marasa, B. S., Zhang, Z., Chopra, A. K. and Rao, V. B. (2013). In vitro and in vivo delivery of genes and proteins using the bacteriophage T4 DNA packaging machine. Proc Natl Acad Sci U S A 110(15): 5846-5851.
- Tao, P., Zhu, J., Mahalingam, M., Batra, H. and Rao, V. B. (2018). Bacteriophage T4 nanoparticles for vaccine delivery against infectious diseases. Adv Drug Deliv Rev 145: 57-72.
- Wang, C., Sun, X., Cheng, L., Yin, S., Yang, G., Li, Y. and Liu, Z. (2014). Multifunctional theranostic red blood cells for magnetic-field-enhanced in vivo combination therapy of cancer. Adv Mater 26(28): 4794-4802.
- Yoo, J. W., Irvine, D. J., Discher, D. E. and Mitragotri, S. (2011). Bio-inspired, bioengineered and biomimetic drug delivery carriers. Nat Rev Drug Discov 10(7): 521-535.
- Zhang, Z., Kottadiel, V. I., Vafabakhsh, R., Dai, L., Chemla, Y. R., Ha, T. and Rao, V. B. (2011). A promiscuous DNA packaging machine from bacteriophage T4. PLoS Biol 9(2): e1000592.
- Zhu, J., Tao, P., Mahalingam, M., Sha, J., Kilgore, P., Chopra, A. K. and Rao, V. (2019). A prokaryotic-eukaryotic hybrid viral vector for delivery of large cargos of genes and proteins into human cells. Sci Adv 5(8): eaax0064.
Article Information
Copyright
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Zhu, J., Tao, P., Mahalingam, M. and Rao, V. (2020). Preparation of a Bacteriophage T4-based Prokaryotic-eukaryotic Hybrid Viral Vector for Delivery of Large Cargos of Genes and Proteins into Human Cells. Bio-protocol 10(7): e3573. DOI: 10.21769/BioProtoc.3573.
- Zhu, J., Tao, P., Mahalingam, M., Sha, J., Kilgore, P., Chopra, A. K. and Rao, V. (2019). A prokaryotic-eukaryotic hybrid viral vector for delivery of large cargos of genes and proteins into human cells. Sci Adv 5(8): eaax0064.
Category
Molecular Biology > DNA > DNA recombination
Molecular Biology > DNA > Gene expression
Molecular Biology > Nanoparticle > Plan-derived nanoparticles
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








