- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Method for Measuring Mucociliary Clearance and Cilia-generated Flow in Mice by ex vivo Imaging
Published: Vol 10, Iss 6, Mar 20, 2020 DOI: 10.21769/BioProtoc.3554 Views: 4964
Reviewed by: Andrea PuharValerian DORMOYAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

A Novel Optimized Silver Nitrate Staining Method for Visualizing and Quantifying the Osteocyte Lacuno-Canalicular System (LCS)
Jinlian Wu [...] Libo Wang
Apr 20, 2025 1520 Views

Standardized Flow Cytometry Method for Absolute Counting of Intraepithelial Lymphocytes in the Intestinal Mucosa Using TruCountTM Beads
Corentin Joulain [...] Salima Hacein-Bey-Abina
May 5, 2025 2189 Views

PBMC-Humanized Mouse Model for Multiple Sclerosis: Studying Immune Changes and CNS Involvement
Anastasia Dagkonaki [...] Lesley Probert
May 20, 2025 3976 Views
Abstract
Ex vivo biophysical measurements provide valuable insights into understanding both physiological and pathogenic processes. One critical physiological mechanism that is regulated by these biophysical properties is cilia-generated flow that mediates mucociliary clearance, which is known to provide protection against foreign particles and pathogens in the upper airway. To measure ciliary clearance, several techniques have been implemented, including the use of radiolabeled particles and imaging with single-photon emission computerized tomography (SPECT) methods. Although non-invasive, these tests require the use of specialized equipment, limiting widespread use. Here we describe a method of ex vivo imaging of cilia-generated flow, adapted from previously reported methods, to make it more accessible and higher throughput for researchers. We excise trachea from mice quickly after euthanasia, cut it longitudinally and place it in an inhouse made slide. We apply fluorescent particles to measure particle movement under a fluorescent microscope, followed by analysis with ImageJ, allowing calculation of fluid flow generated by cilia under different conditions. This method enables ex vivo measurements in tissue with minimal investment or special equipment, giving opportunity to investigate and discover important biophysical properties associated with ciliary movement of the trachea in physiology and disease.
Keywords: CiliaBackground
The composition of diverse biophysical properties, each aiding a specified function, make up the human body. A key example of this is the generation of flow in many different organ systems, including the central nervous system (Olstad et al., 2019), reproductive tracts (Afzelius et al., 1978; Yuan et al., 2019) and the pulmonary system (Satir and Sleigh, 1990), made by specialized ciliated cells. Understanding how the environment or external stimuli affect these processes can give us insights into disease. In our recent study (Kudo et al., 2019), we explored how humidity affects influenza infections in a mouse model. By using this method of ex-vivo measurement of cilia-generated flow in mice, we were able to visually measure how ambient humidity altered the rate of virus clearance (Figures 1 and 2). Previous studies have used radiolabeled particles to measure mucociliary clearance using a noninvasive method (Hua et al., 2010; Bustamante-Marin and Ostrowski, 2017). However, this requires the use of radioactive materials and SPECT or other machines to measure radioactivity that may not be readily available. Measurements like this can be made more accessible using ex vivo imaging of particles in tissue (Nance et al., 2012; Francis and Lo, 2013; Mastorakos et al., 2015 and 2016). Here, we describe a detailed method of measuring the flow generated by cilia in upper airway tissue, adapted from Francis and Lo (2013). Our method is easy to perform and allows for measurement of large numbers of mice at a relatively low cost. This protocol can also be performed in a short time, allowing for the preservation of biological differences and measurement in a large number of animals. Finally, it can be adopted for other organ systems or tissues, measuring various biophysical properties, as shown by Nance et al. (2012), in identifying what criteria drug loaded particles must meet to navigate through brain parenchyma. Altogether, this protocol will provide researchers with an easy to follow, step-by-step methodology to measure properties of biological tissues ex vivo (Figure 3).
Materials and Reagents
- Superfrost Plus Gold Slides (Thermo Fisher Scientific, catalog number: FT4981GLPLUS ). Brand is not critical
- Square Cover Slips (Thermo Fisher Scientific, catalog number: 18X18-1 ). Brand is not critical
- Electrical tape (3M Safety, Scotch, Purchase in Amazon). Brand is not critical
- FluoSpheres carboxylate, 0.2 μm crimson, 625/645 (Life Technologies, catalog number: F8806 ) Alternate sizes/chemistry can be used depending on experimental design, refer to background above
- 6-well plate (Fisher Scientific, Corning). Brand is not critical, does not have to be tissue culture quality
- Mice
- PBS (Millipore Sigma, catalog number: D8537 ). Brand is not critical
- Super glue (Loctite, Purchase in Amazon). Brand is not critical
- 1:1,000 volume/volume of FluoSpheres carboxylate in PBS (see Recipes)
Equipment
- Dissection scissors (Fine Science Tools, catalog number: 14061-10 ) Brand is not critical
- Electric Razor (Purchase from Amazon) Brand is not critical
- Hair removal product, Nair (Purchase from Amazon) Brand is not critical
- Forceps (Fine Science Tools) Brand is not critical
- Micro dissection scissors (Fine Science Tools) Brand is not critical
- Scalpel (Purchase from Amazon) Optional and brand is not critical
- Dissection microscope (Laxco, LMS-Z200) Optional and brand is not critical
- Bruker Opterra Swept Field Microscope (Bruker) Brand is not critical, microscope must be equipped with high-speed fluorescent camera (Photometrics Evolve Delta EMCCD camera)
Software
- Obtaining images Prairie View 5.4 (Bruker) Brand is not critical, any software associated with microscope will work
- Analysis Software ImageJ (Directionality, Mtrack2, Manual Tracking, NIH, https://imagej.nih.gov/ij/) Other software can be used for analysis
Procedure
- Prepare slides
- Make a stack of electrical tape to a thickness of ~1.5 mm (7 pieces of tape) (Figures 1A-1B).
Note: The thickness of the tape here can vary depending on the thickness of tissue you are interested in working with, with the thickness corresponding to the tissue touching both the bottom of the slide and the coverslip without distorting the tissue significantly. However, it is important to keep the thickness consistent between samples, as thickness can affect your imaging later on. - Cut a small section using a scalpel to allow for housing of the trachea (Figure 1C).
- Put the tape on a slide to make a slide with a housing chamber usable with most microscopes (Figure 1D).
Note: Slides can be prepared in large numbers easily before experiment. Do this to speed up throughput of experiments.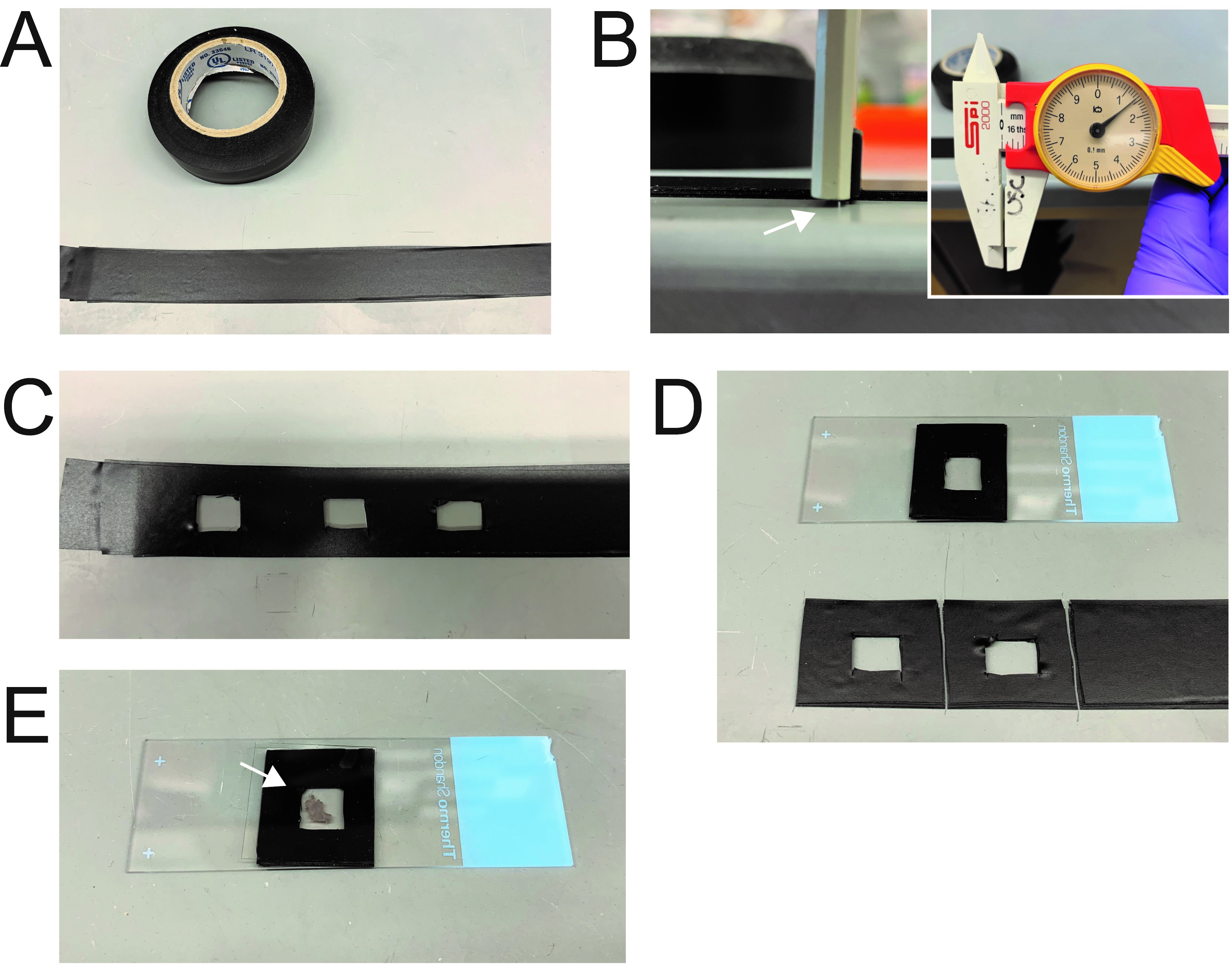
Figure 1. Preparation of chamber slides for measurement of mucociliary clearance. A-B. Electrical tape was stacked on top of each other to make a stack equivalent to ~1.5 mm (white arrow). C. Sections were cut in the center of the tape to create a chamber for tissue. D. Tape was attached to slide for imaging. E. Example of Tracheal tissue that was cut longitudinally and placed in the chamber with cover slip.
- Make a stack of electrical tape to a thickness of ~1.5 mm (7 pieces of tape) (Figures 1A-1B).
- Dissection of trachea (Time sensitive)
The following procedure, till the image acquisition should be performed uninterrupted for each mouse (rather than extracting all tracheae for the experiments) and quickly for best results.- Euthanize mice in accordance with local institutional animal care and use committee (IACUC) policies and protocol.
- Immobilize mouse using a dissection board or by taping mouse onto a hard surface. Immediately expose neck skin using a hair removal product of your choice (Nair, Electric Razor), make a ~3 cm incision from the chest to nose, and move salivary glands aside to expose trachea of mice.
- Clean connective tissue associated with trachea and make transverse cut on the superior end of the trachea. Keep trachea taut by holding this side with forceps.
- Cut inferior end of trachea.
- Immediately transfer to 6 well plate with ~1 ml PBS.
- Cut longitudinally through trachea using the micro dissection scissors.
- Immediately continue onto next step.
- Mounting trachea
- Prepare a solution of 1:1,000 of FluoSpheres carboxylate in PBS.
Note: Prepare large batches for use of your entire experiment. Vortex initial solution well, and vortex the mix well between each use. - Place trachea from Procedure B into slide from Procedure A with the lumen side facing up.
Note: Having the tape height match the tissue thickness is critical, as this allows the coverslip to prevent the trachea from refolding during imaging. - Place ~250 μl of 1:1,000 FluoSphere solution in the chamber, place a few drops of super glue on edges of tape and coverslip (Figure 1E).
Note: Take precaution not to put excess liquid in chamber and to not put excess glue on tape. Aim for having enough liquid to submerge sample, or fully covering the center, without it spilling over. Mixing of the liquid submerging the sample with the super glue will result in unreliable results. Optimize with practice slides to get exact volume needed for each setup. - Immediately continue onto next step.
- Prepare a solution of 1:1,000 of FluoSpheres carboxylate in PBS.
- Imaging
- Microscope should be turned on and set into the correct position. Use a slide with particle solution but no organ as a positive control (refer to Video 1).
Note: Imaging was performed under room temperature. However, experiments can be performed under different temperatures if your microscope setup allows for different temperature settings.Video 1. Example of particles moving in free solution in a brownian motion - Place slide into position.
- *Important* Change focus until you find a plane where particles seem to be static. This is the plane of the tissue, where particles are attached/engulfed to the mucus/cells on the surface. From there move focus up a set amount for each image. This is important because the measurement of the flow generated by cilia will change as a function of the distance away from the surface of the cilia. Thus, to generate data that can be compared between samples, this distance must be kept constant (refer to Video 2). In our experiments, we used a distance of ~1 μm distance from the surface of the cilia.Video 2. Example of particles moving with directionality due to cilia generated flow
- Acquire image at speeds at or above 100 frames per second (we acquired at 100 fps for 30 s). Images were taken with microscope settings; Image Size: 512 μm x 512 μm, FOV: 204.2 μm x 204.2 μm, and Pixel Size 0.399 μm x 0.399 μm.
Note: The two imaging lasers in our experiments were 488 nm and 561 nm, with the emission filters optimized for GFP and RFP. The two cameras on the microscope are 512 μm x 512 μm Photometrics Evolve Delta EMCCD cameras. The Laser is scanned with either a slit or an array of 32 pinholes, which is designed to increase scan speed, which is 67 fps max speed at full frame, with smaller ROIs allowing for much faster speeds. - It is critical that these images are taken as quickly as possible; in our hands, all images were taken within 5 min of dissection, and saw decreased viability of tissue after 10 min.
Note: The key component to this step is feeling comfortable with the procedure and decreasing the time between dissection and imaging. Practice to get a good flow going, or work in pairs to have one person dissect while another sets up the microscope in order to set up an optimal imaging condition. As a positive control of cilia movement regulation, mice housed in higher ambient humidity (~50%) will show higher cilia particle velocity (Kudo et al., 2019). As a negative control, mouse housed in lower ambient humidity can be used, or trachea that has be excised for a prolonged time period (> 10 min) can be used.
- Microscope should be turned on and set into the correct position. Use a slide with particle solution but no organ as a positive control (refer to Video 1).
Data analysis
- Load data into FIJI/ImageJ using File > Import > Image sequence.
- Analyze particle velocity using Plugins > Tracking > MTrack2 (https://imagej.net/MTrack2).
Note: Use imageJ settings Image > Adjust > Brightness/Contrast to ensure a high signal to noise ratio. This will allow for the MTrack2 program to identify the particles better. - Measure single particle trajectory by using Plugins > Tracking > Manual Tracking (https://imagej.nih.gov/ij/plugins/track/track.html) (Figure 2).
Note: This function will allow for coordinates of a single particle to be recorded. This can be then graphed using a graphing software to show the entire trajectory of a particle over a given time. - Calculate directionality of flow using Analyze > Directionality (Figure 3).
- Export video (Videos 1 and 2) versions of particle flow using File > Save As > AVI.
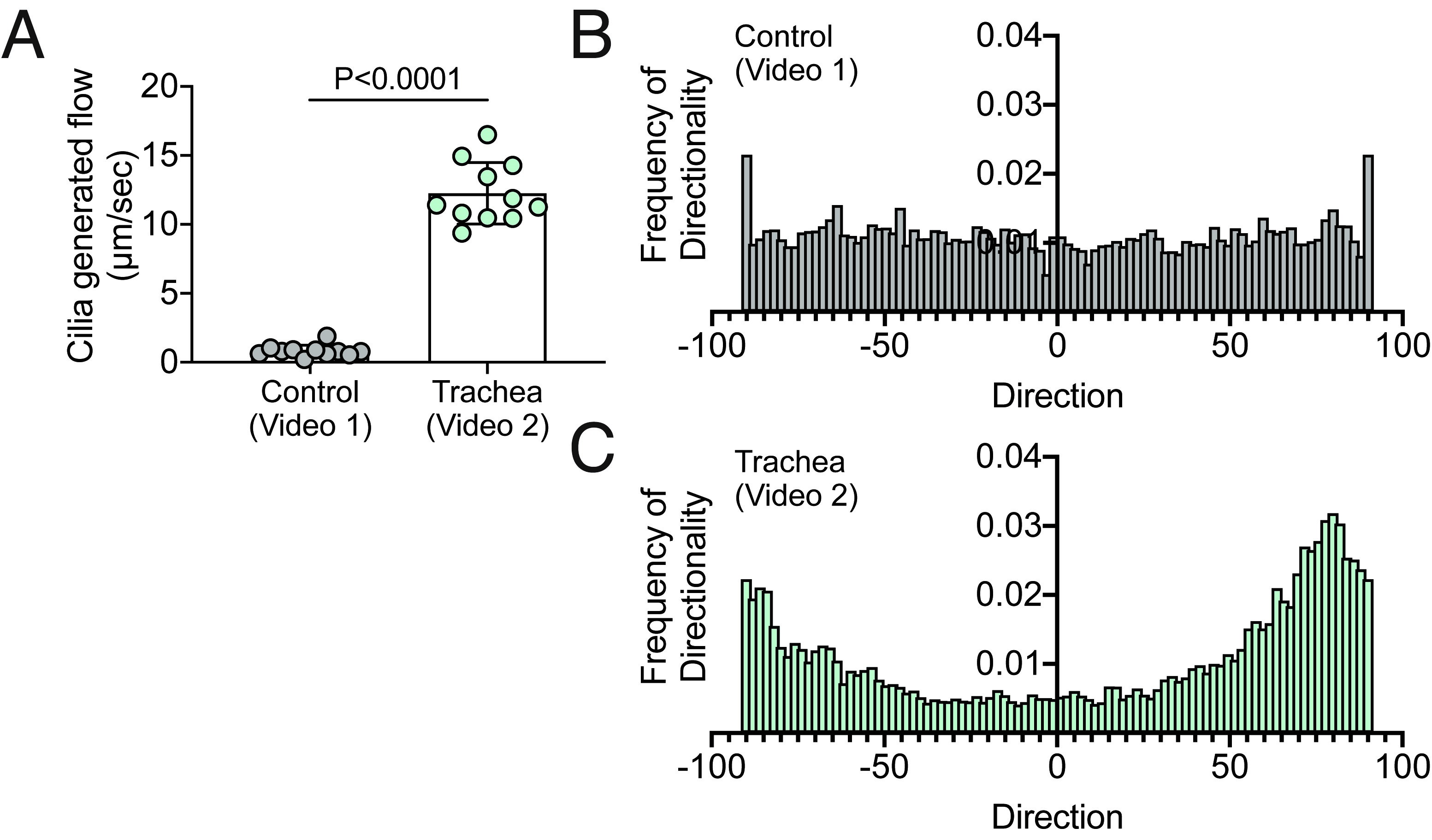
Figure 2. Cilia generated flow and directionality analysis of Videos 1 and 2. A. Quantification of cilia generated flow in Videos 1 and 2 using Data analysis section of the protocol. B-C. Quantification of frequency of directionality of Videos 1 and 2.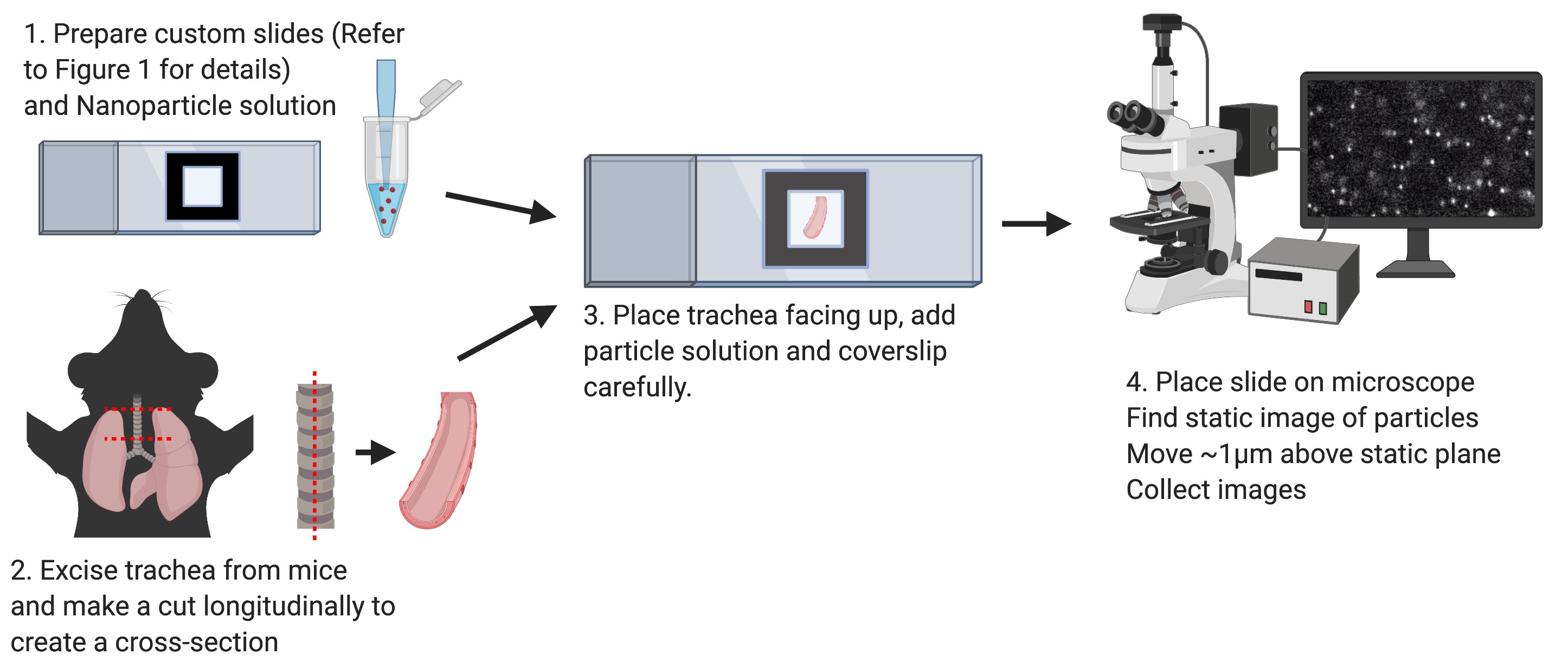
Figure 3. Schematic of principal steps in cilia generated flow measurement
Recipes
- 1:1,000 volume/volume of FluoSpheres carboxylate in PBS
Solution should be stored in the dark
We made fresh solution each day and equilibrated to room temperature before use
Acknowledgments
We thank Melissa Linehan for technical and logistical assistance. We thank Al Mennone with help setting up the microscope for these experiments and Eriko Kudo for experiments that were performed that led to the original research paper. Graphical illustrations were made with Biorender.com. This work was supported in part by the Howard Hughes Medical Institute (A.I.), a gift from the Condair Group and National Institutes of Health Grants T32GM007205 (Medical Scientist Training Program training grant to E.S.). The original research paper for this protocol can be found at Kudo et al. (2019).
Competing interests
The authors have no financial and non-financial competing interests regarding the protocols described in this manuscript.
Ethics
No human subjects were used in this study. All animal work was approved under Yale IACUC protocol 2018-10365.
References
- Afzelius, B. A., Camner, P. and Mossberg, B. (1978). On the function of cilia in the female reproductive tract. Fertil Steril 29(1): 72-74.
- Bustamante-Marin, X. M. and Ostrowski, L. E. (2017). Cilia and mucociliary clearance. Cold Spring Harb Perspect Biol 9(4).
- Francis, R. and Lo, C. (2013). Ex vivo method for high resolution imaging of cilia motility in rodent airway epithelia. J Vis Exp(78).
- Hua, X., Zeman, K. L., Zhou, B., Hua, Q., Senior, B. A., Tilley, S. L. and Bennett, W. D. (2010). Noninvasive real-time measurement of nasal mucociliary clearance in mice by pinhole gamma scintigraphy. J Appl Physiol (1985) 108(1): 189-196.
- Kudo, E., Song, E., Yockey, L. J., Rakib, T., Wong, P. W., Homer, R. J. and Iwasaki, A. (2019). Low ambient humidity impairs barrier function and innate resistance against influenza infection. Proc Natl Acad Sci U S A 116(22): 10905-10910.
- Mastorakos, P., da Silva, A. L., Chisholm, J., Song, E., Choi, W. K., Boyle, M. P., Morales, M. M., Hanes, J. and Suk, J. S. (2015). Highly compacted biodegradable DNA nanoparticles capable of overcoming the mucus barrier for inhaled lung gene therapy. Proc Natl Acad Sci U S A 112(28): 8720-8725.
- Mastorakos, P., Song, E., Zhang, C., Berry, S., Park, H. W., Kim, Y. E., Park, J. S., Lee, S., Suk, J. S. and Hanes, J. (2016). Biodegradable DNA Nanoparticles that Provide Widespread Gene Delivery in the Brain. Small 12(5): 678-685.
- Nance, E. A., Woodworth, G. F., Sailor, K. A., Shih, T. Y., Xu, Q., Swaminathan, G., Xiang, D., Eberhart, C. and Hanes, J. (2012). A dense poly(ethylene glycol) coating improves penetration of large polymeric nanoparticles within brain tissue. Sci Transl Med 4(149): 149ra119.
- Olstad, E. W., Ringers, C., Hansen, J. N., Wens, A., Brandt, C., Wachten, D., Yaksi, E. and Jurisch-Yaksi, N. (2019). Ciliary beating compartmentalizes cerebrospinal fluid flow in the brain and regulates ventricular development. Curr Biol 29(2): 229-241 e226.
- Satir, P. and Sleigh, M. A. (1990). The physiology of cilia and mucociliary interactions. Annu Rev Physiol 52: 137-155.
- Yuan, S., Liu, Y., Peng, H., Tang, C., Hennig, G. W., Wang, Z., Wang, L., Yu, T., Klukovich, R., Zhang, Y., Zheng, H., Xu, C., Wu, J., Hess, R. A. and Yan, W. (2019). Motile cilia of the male reproductive system require miR-34/miR-449 for development and function to generate luminal turbulence. Proc Natl Acad Sci U S A 116(9): 3584-3593.
Article Information
Copyright
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Song, E. and Iwasaki, A. (2020). Method for Measuring Mucociliary Clearance and Cilia-generated Flow in Mice by ex vivo Imaging. Bio-protocol 10(6): e3554. DOI: 10.21769/BioProtoc.3554.
Category
Immunology > Mucosal immunology > Epithelium
Immunology > Animal model > Mouse
Cell Biology > Tissue analysis > Tissue imaging
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










