- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Quantification of Protein Enrichment at Plasmodesmata
Published: Vol 10, Iss 5, Mar 5, 2020 DOI: 10.21769/BioProtoc.3545 Views: 5500
Reviewed by: Ali ParsaeimehrTohir BozorovAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

A Detailed Guide to Recording and Analyzing Arabidopsis thaliana Leaf Surface Potential Dynamics Elicited by Mechanical Wounding
Fatiha Atanjaoui [...] Michael M. Wudick
Apr 5, 2025 1442 Views

Near-Infrared Autofluorescence Imaging of Nuclei in Living Plant Roots
Akira Yoshinari and Masayoshi Nakamura
Apr 20, 2025 2122 Views

Live-Cell Monitoring of Piecemeal Chloroplast Autophagy
Masanori Izumi [...] Shinya Hagihara
Nov 5, 2025 1688 Views
Abstract
Intercellular communication plays a crucial role in the establishment of multicellular organisms by organizing and coordinating growth, development and defence responses. In plants, cell-to-cell communication takes place through nanometric membrane channels called plasmodesmata (PD). Understanding how PD dictate cellular connectivity greatly depends on a comprehensive knowledge of the molecular composition and the functional characterization of PD components. While proteomic and genetic approaches have been crucial to identify PD-associated proteins, in vivo fluorescence microscopy combined with fluorescent protein tagging is equally crucial to visualise the subcellular localisation of a protein of interest and gain knowledge about their dynamic behaviour. In this protocol we describe in detail a robust method for quantifying the degree of association of a given protein with PD, through ratiometric fluorescent intensity using confocal microscopy. Although developed for N. benthamiana and Arabidopsis, this protocol can be adapted to other plant species.
Keywords: PlasmodesmataBackground
Currently, confirmation of protein localization to PD by confocal imaging is based primarily on two different approaches. On the one hand, the molecular composition of the cell wall surrounding PD differs. While the cell wall is highly enriched in cellulose, the environment near the PD is enriched in callose, a beta (1-3) glucan polymer that can easily and specifically be stained with fluorophore Aniline Blue. This staining presents the considerable advantage of being used as a PD marker without crossed lines. On the other hand, proteomic studies have identified specific PD proteins and led to their characterization (Faulkner et al., 2005; Fernandez-Calvino et al., 2011; Grison et al., 2015; Brault et al., 2019). These proteins can be used as PD markers in subsequent studies when tagged with fluorescent proteins in transient or stable expression in plants (Thomas et al., 2008; Simpson et al., 2009). Note that PD proteins can be exclusively associated with PD but can also present a dual localization within different cellular compartments as for Synaptotagmin1 (SYT1), Multiple C2 domains and Transmembrane regions Protein 4 (MCTP4) which associate with both the Endoplasmic Reticulum and PD (Levy et al., 2015; Brault et al., 2019). Since PD are dynamic structures responding to developmental and environmental cues, their molecular constituents may vary conditionally (Benitez-Alfonso et al., 2013; Stahl et al., 2013; Han et al., 2014; Sager and Lee, 2014; Otero et al., 2016; Stahl and Faulkner, 2016). Thus, Receptor Like Kinases (RLKs), such as the Leucine-Rich-Repeat RLKs Qian Shou Kinase 1 (QSK1) and Inflorescence Meristem Kinase 2 (IMK2) or the Cystein-Rich Receptor Kinase 2 (CRK2), are able to dynamically relocate to PD upon osmotic stress conditions (Grison et al., 2019; Hunter et al., 2019). The study of the dynamic localization of protein in vivo requires the development of quantification methodologies. Using confocal microscopy, we developed a ratiometric calculations to evaluate the PD enrichment of a given protein using PD markers, hereinafter referred as “PD Index”. The PD Index can be used for co-localization experiments but also as reference points for characterizing mutants, drugs or growing conditions that could modify the degree of proteins PD association (Perraki et al., 2018; Brault et al., 2019; Grison et al., 2019).
Materials and Reagents
- Syringe without needle (1 ml, Dutscher, catalog number: 8SS01H1 )
- Razorblade (19 x 38 x 0.27 mm, Dutscher, catalog number: 320529 )
- Slides (76 x 26 mm, Dutscher, catalog number: 0 6962 )
- Coverslips (22 x 32 mm, Dutscher, catalog number: 100034M )
- Tweezer (Pince à écharde forme pointue, Dutscher, catalog number: 711197 )
- Plant material (see Procedure A)
- Aniline Blue (Biosupplies Australia, catalog number: 100.1 ), storage at 4 °C
Note: The Aniline Blue stock solution is 1 mg/ml in water. The Aniline Blue working solution is 0.025 mg/ml. Do not use higher concentration of aniline bleu when the protein of interest is GFP tagged otherwise the Aniline Blue signal will crosstalk with the GFP signal. - Distilled water
- Luria and Bertani medium (LB broth Miller, Sigma-Aldrich, catalog number: L3152 )
- Sucrose (Sigma-Aldrich, catalog number: S7903 )
Equipment
- Confocal Microscope: plant imaging was performed using a ZEISS microscope (ZEISS, model: LSM880 )
- 28 °C Shaking Incubator (Dutscher, MaxQ 4000, catalog number: 0 78381 )
- Centrifuge (Dutscher, Spectrafuge 6 C for 10 ml tubes, catalog number: 0 96610 )
Software
- FIJI (https://imagej.net/Fiji) (Schindelin et al., 2012)
- R (https://www.r-project.org) (R Core Development Team, 2015)
Procedure
- Plant material preparation
In Arabidopsis seedlings- Aniline Blue staining
This method allows PD staining in the cotyledon and the hypocotyl, the aniline blue staining in roots gives a resolution and staining efficiency that we find difficult to combine with PD Index calculation.
Note: For roots, we advise the users to do immunolocalization in whole mount roots using monoclonal antibody: Biosupplies Australia, (1-3)-beta-glucan-directed monoclonal, catalogue number 400-2 and the protocol described in Boutté and Grebe, 2014.- Grow Arabidopsis seedlings during 4 to 6 days on ½ MS 1% sucrose agar plate under 16/8 h day/night photoperiodic condition (150 μE/m2/s, 22 °C).
- Take 0.2 ml of Aniline blue solution at a concentration of 0.025 mg/ml in water with the 1 ml syringe.
Note: Abiotic or biotic stress conditions can be tested to compare the PD Index of the protein of interest, in that case the aniline blue can be diluted in water supplemented with different molecules (such as NaCl, Mannitol, …) or can be apply before the aniline blue staining (such as viral infection where the plant is infected few days before aniline blue infiltration). Note that leaves or seedlings cannot be infiltrated twice. - Remove the air from the syringe.
- Carefully take the seedling with tweezers.
- Gently push the seedling into the syringe, so that the aerial parts of the seedling (i.e., the cotyledons and hypocotyl) are immersed in the solution (Figures 1A-1D). The seedling’s root should protrude from the syringe in order to be able to extract the seedling from the syringe after infiltration.
- Position your finger at the extremity of the syringe and slowly pull the piston out of the syringe and reach the 0.21 graduation, bubbles should appear at the surface of the cotyledons (Figures 1E-1H).
- Count to 5 and then release the piston very slowly (Figures 1I).
Note: We recommended the users to be extremely delicate and gentle at both steps (f and g) otherwise the cells will explode under the pressure and it will lead to a general blurry blue coloration. - Carefully remove the seedling from the syringe by grasping the root of it with tweezers (Figures 1J-1K).
- Place the seedling on a slide in a drop of water, and gently mount the coverslip.
- Immediately proceed to the acquisition under the confocal microscope.
Note: Aniline Blue bleaches rapidly. Direct observation should be done with a low power of the mercury lamp to avoid the bleaching of the sample. In confocal mode, please use a 405 nm laser power as low as possible for the same reason. With the ZEISS 880 confocal device, the laser power is generally set around 0.2 to 5%, but of course this may vary depending on the microscope used. Aniline blue staining can be visualised immediately and is stable during 10 to 15 min.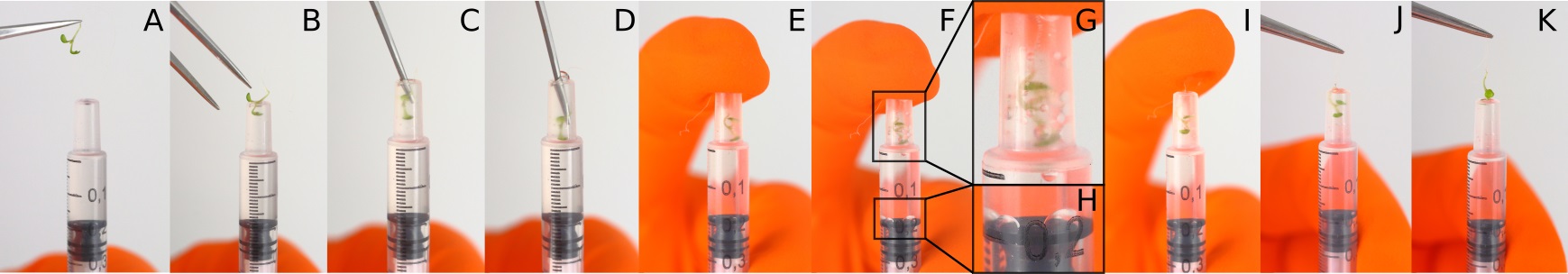
Figure 1. Arabidopsis seedling infiltration. Illustration of the different steps of an Arabidopsis seedling infiltration using a 1 ml syringe.
- Using PD proteins as PD marker
In addition to aniline blue, fluorescently tagged PD proteins such as Plasmodesmata Located Protein 1 (PDLP1) or Plasmodesmata Callose Binding Protein 1 (PDCB1) can be used as PD markers for the PD Index calculation both in transient and stable expression (Thomas et al., 2008; Simpson et al., 2009). However, we highly recommend crossing the Arabidosis lines expressing the protein of interest with the available Arabidopsis lines expressing a fluorescent tagged PD marker protein.
Notes:- Over expression of PD associated proteins can lead to callose deposition at PD.
- Transient expression of fluorescent tagged PD protein markers in Arabidopsis seedlings may be used but the efficiency rate of transformation is low. We do not recommend this method.
- Aniline Blue staining
- Three days before imaging, infiltrate Nicotiana benthamiana leaves only with agrobacteria previously electroporated with the relevant binary plasmid of the protein of interest (as described in Step A1a of In Arabidopsis seedlings).
- Take a small volume, around 0.2 ml, of aniline blue at a concentration of 0.025 mg/ml in water in a 1 ml syringe.
- Gently return the leaf to show its abaxial side.
- Apply the syringe on the leaf and position your finger at the same location on the other side in order to block the syringe (Figures 2A-2B).
Note: Avoid infiltration in veins area of the leaf, the area to infiltrate should be as flat as possible. Also avoid the puncture site from the previous Agrobacterium infiltration. - Apply a small pressure on the leaf with the syringe and push slowly the piston in order to infiltrate (Figures 2C-2F).
Note: When the liquid penetrates and migrates in the leaf, a darker area should appear and expend around the syringe. If the pressure applied is correct, the infiltration should be smooth, without resistance nor loss of liquid, and without wounding the surface of the leaf. - Do not infiltrate the whole leaf, a small area of 1 cm2 is sufficient.
- With a sharp razor blade, cut the infiltrated area of the leaf.
- Place the sample on a mounting slide, abaxial side of the leaf facing up.
- Add a drop of water on the sample.
- Cover it with the coverslip.
- Immediately proceed to the acquisition under the confocal microscope.
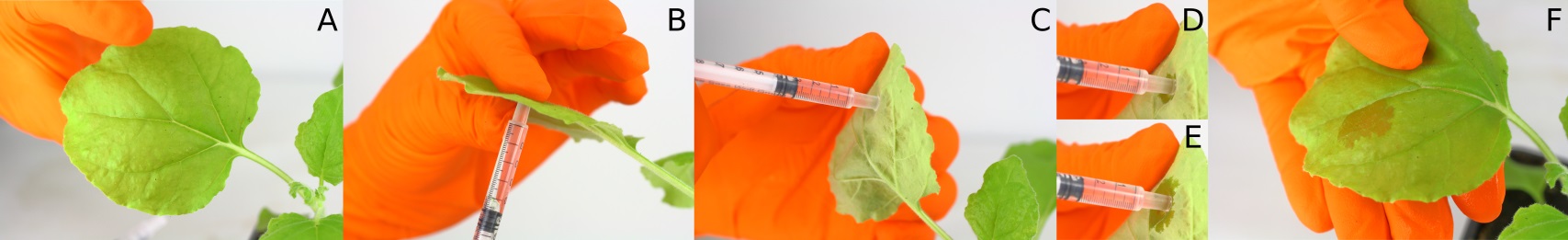
Figure 2. Nicotiana benthamiana leave infiltration. Illustration of the different steps (A-F) of Nicotiana benthamiana leaf infiltration using a 1 ml syringe.
- Using PD proteins as PD marker
- Grow Agrobacterium previously electroporated with the relevant binary plasmids of the protein of interest and of the PD marker (Table 1) in liquid Luria and Bertani medium with appropriate antibiotics, at 28 °C and 250 rpm, for 1 day.
- Perform a 1/10 dilution of each culture and grow again at 28 °C and 250 rpm until the culture reach an OD600 of about 0.8
- Centrifuge the culture at 3,500 x g, discard carefully the supernatant and resuspend in water for a final OD600 of 0.3.
- Mix 1:1 volume of both the agrobacteria cultures transformed with the protein of interest and with the PD marker.
- Use 5 to 6 leaves stage plant and make a very small puncture on the abaxial side of each selected leaf. Avoid the leaf veins.
- Apply a 1 ml syringe containing the agrobacteria mix in water against the leaf at the puncture site
- Position your finger on the other side to block the syringe and gently push the piston while applying a small pressure on the leaf so the liquid can infiltrate. Here, it is advantageous to infiltrate a large portion of the leaf.
- Place the plant in the appropriate culture room for another 3 days for protein expression.
- Using a razor blade, cut a square of approximately 1 cm2 in the infiltrated area of the leaf.
- Place the sample on a mounting slide, abaxial side of the leaf facing up.
- Add a drop of water on the sample.
- Cover it with the coverslip.
- Immediately proceed to the acquisition under the confocal microscope.
- Aniline Blue staining
- Confocal acquisition
- The use of a water immersion objective 63x (NA ≥1.4) for observation is recommended to have the same refraction index between immersion and mounting. If water immersion objective is not available, the use of an oil-immersion objective is still possible.
- The pinhole value needs to be kept at airy 1 to ensure a focal plane as accurate as possible.
- The excitation wavelength and the spectral acquisition windows should be adjusted according to the fluorescent proteins chosen in your experiment (Table 1).
- Laser power, photomultiplier (PMT) and photomultiplier offset should be set such that the acquired signals are not saturated. Between experiments and in a same experiment the setting parameters of the PD marker can be modified to obtain the better signal as possible without bleaching. In a same experiment the fluorescent tagged protein of interest channel setting must be kept identical while comparing different mutants or conditions. Between experiments we also recommend keeping the same settings for the fluorescently-tagged protein of interest. However, the PD Index is a ratiometric calculation between Region Of Interest (ROI) in a same picture so changes on the fluorescent protein acquisition setting may be acceptable if really needed.
- Line average can be applied during image acquisition, the same line average should be kept in all experiments.
- During image acquisition, the sequential scanning is preferable compared to simultaneous scanning. It is important to control that the excitation wavelength used for one fluorescent protein does not excite the other one, i.e., ensure there is no crosstalk between the PD marker/Aniline blue and the tag of the protein of interest (Table 1).
Notes:- The Aniline Blue stock solution is 1 mg/ml in water. The Aniline Blue working solution is 0.025 mg/ml. Do not use higher concentration of aniline bleu when the protein of interest is GFP tagged otherwise the Aniline Blue signal will crosstalk with the GFP signal.
- To avoid crosstalk of fluorescence between channels, it is necessary to perform independent and combined excitation and detection at the excitation and emission wavelength of the individual fluorochrome or fluorescent protein, respectively. Control samples labeled only with a single fluorescent protein/PD marker should be prepared in order to verify that the laser excitation wavelength used to excite one fluorochrome does not excite the other.
Table 1. Fluorescence excitation and emission maximum of Aniline Blue and commonly used fluorescent proteins (FP: Fluorescent Protein or Dye; Ex: Excitation wavelength in nm; Em: Emission wavelength in nm)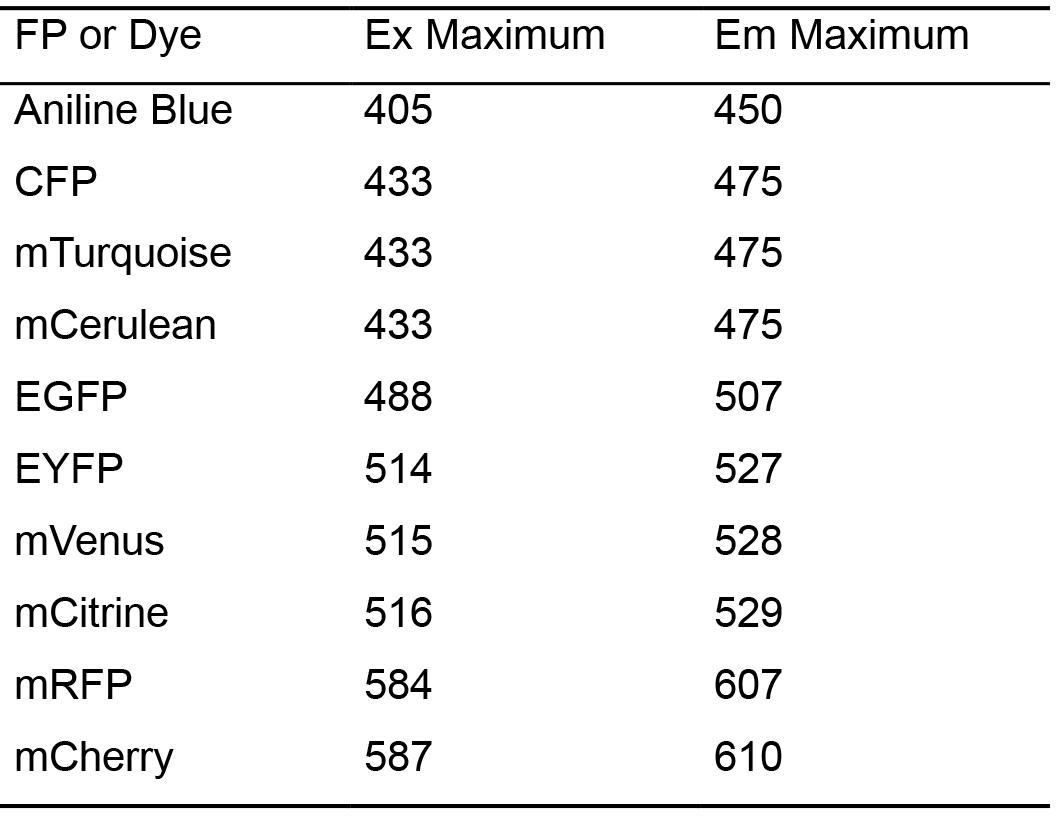
Data analysis
- Data collection using Fiji
- Open the confocal image with Fiji. Do not split channels.
- Open ROI Manager (Analyze > Tools > ROI Manager).
- Use the circle selection tool from main panel and draw a circle on the PD marker channel that is slightly smaller than the size of the PD marker signal (Figure 3A).
Note: Only sharp callose signal should be selected as PD ROI. Do not select ROI either for PD ROI and for PM ROI when the “spot” in the aniline blue channel is blurry. - Click “add” on the ROI Manager once the circle is well positioned and selected. A new line should appear in the ROI Manager.
- Move the circle, without modifying it, to another signal and click “add” again.
- Repeat Step A4 until you have enough ROI (ROI; best between 5 and 10).
- Change the view to the other channel (the protein of interest fluorescence channel).
- Using the same circle resume from step A5 and add regions that do not overlap with the AB signals but are at the cell periphery (Figure 3B).
- Verify the measurement parameters by going to Analyze > Set parameters. Tick the “Mean gray value” box.
- Verify that the channel is still on the protein of interest. Then, on the ROI Manager, click on “Measure”. A new window appears with the values.
- Select and copy the values.
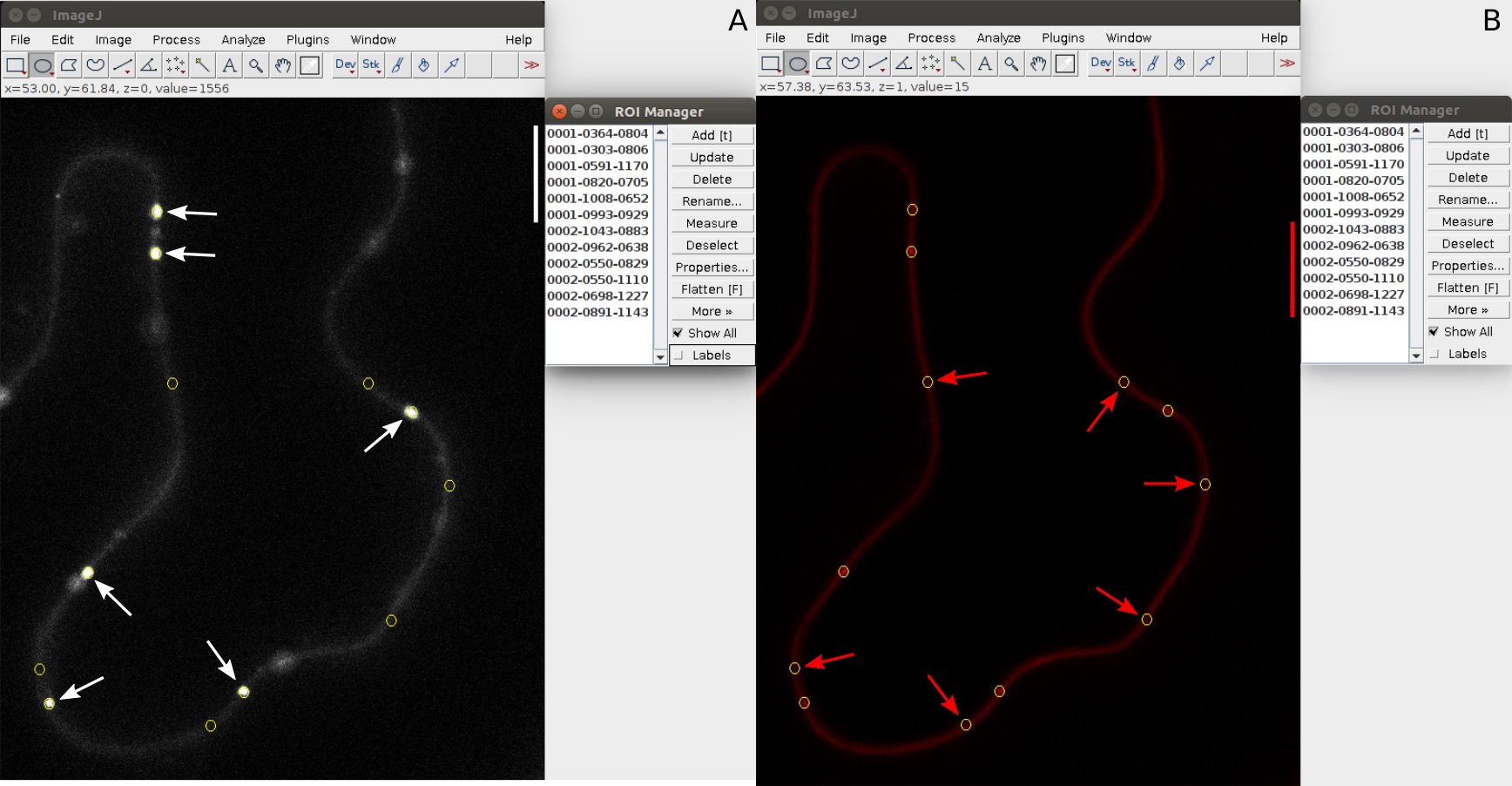
Figure 3. ROI selection for PD Index calculation. A. The PD ROI are selected in the PD marker channel (white arrows). B. The PM ROI are selected in the protein of interest fluorescence channel (red arrows). Verify that the ROI selected in the protein of interest fluorescence channel do not correspond to a PD in the PD marker channel.
- PD index calculation using Excel
- On an Excel sheet, paste the values copied from Fiji (ImageJ)
- Calculate the mean of the values from “at PD” ROIs and the mean of the values from “outside PD” ROIs
- Calculate the PD index by doing “At PD ROI mean value/Outside PD ROI mean value”. A PD Index value below or equal to 1 means that there is no specific enrichment nor accumulation of the protein of interest at PD, whereas a PD Index value above 1 is significant from enrichment of a protein at PD.
For statistical analysis the use of R software and the Rcmdr package is recommended. Parametrical tests can be use only when n ≥ 20 and when the sample distribution respect the normal law. Non-parametrical tests are systematically used when n < 20.
Acknowledgments
J.D.P. is funded by a PhD fellowship from the Belgian “Formation à la Recherche dans l'Industrie et l'Agriculture” (FRIA grant no. 1.E.096.18).
This work was supported by the National Agency for Research (Grant ANR-14-CE19-0006-01 to E.M.B), “Osez l’interdisciplinarité” OSEZ-2017-BBRIDGING CNRS program to E.M.B., the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement No 772103-BRIDGING to E.M.B).
Competing interests
The authors declare no competing financial interests.
References
- Benitez-Alfonso, Y., Faulkner, C., Pendle, A., Miyashima, S., Helariutta, Y. and Maule, A. (2013). Symplastic intercellular connectivity regulates lateral root patterning. Dev Cell 26(2): 136-147.
- Boutté, Y. and Grebe, M. (2014). Immunocytochemical fluorescent in situ visualization of proteins in Arabidopsis. Methods Mol Biol 1062: 453-472.
- Brault, M. L., Petit, J. D., Immel, F., Nicolas, W. J., Glavier, M., Brocard, L., Gaston, A., Fouché, M., Hawkins, T. J., Crowet, J. M., Grison, M. S., Germain, V., Rocher, M., Kraner, M., Alva, V., Claverol, S., Paterlini, A., Helariutta, Y., Deleu, M., Lins, L., Tilsner, J. and Bayer, E. M. (2019). Multiple C2 domains and transmembrane region proteins (MCTPs) tether membranes at plasmodesmata. EMBO Rep 20(8): e47182.
- Faulkner, C. R., Blackman, L. M., Cordwell, S. J. and Overall, R. L. (2005). Proteomic identification of putative plasmodesmatal proteins from Chara corallina. Proteomics 5(11): 2866-2875.
- Fernandez-Calvino, L., Faulkner, C., Walshaw, J., Saalbach, G., Bayer, E., Benitez-Alfonso, Y. and Maule, A. (2011). Arabidopsis plasmodesmal proteome. PLoS One 6(4): e18880.
- Grison, M. S., Kirk, P., Brault, M. L., Wu, X. N., Schulze, W. X., Benitez-Alfonso, Y., Immel, F. and Bayer, E. M. (2019). Plasma membrane-associated receptor-like kinases relocalize to plasmodesmata in response to osmotic stress. Plant Physiol 181(1): 142-160.
- Grison, M. S., Brocard, L., Fouillen, L., Nicolas, W., Wewer, V., Dörmann, P., Nacir, H., Benitez-Alfonso, Y., Claverol, S., Germain, V., Boutté, Y., Mongrand, S. and Bayer, E. M. (2015). Specific membrane lipid composition is important for plasmodesmata function in Arabidopsis. Plant Cell 27(4): 1228-1250.
- Han, X., Kumar, D., Chen, H., Wu, S. and Kim, J. Y. (2014). Transcription factor-mediated cell-to-cell signalling in plants. J Exp Bot 65(7): 1737-1749.
- Hunter, K., Kimura, S., Rokka, A., Tran, H. C., Toyota, M., Kukkonen, J. P. and Wrzaczek, M. (2019). CRK2 enhances salt tolerance by regulating callose deposition in connection with PLDα1. Plant Physiol 180(4): 2004-2021.
- Levy, A., Zheng, J. Y. and Lazarowitz, S. G. (2015). Synaptotagmin SYTA forms ER-plasma membrane junctions that are recruited to plasmodesmata for plant virus movement. Curr Biol 25(15): 2018-2025.
- Otero, S., Helariutta, Y. and Benitez-Alfonso, Y. (2016). Symplastic communication in organ formation and tissue patterning. Curr Opin Plant Biol 29: 21-28.
- Perraki, A., Gronnier, J., Gouguet, P., Boudsocq, M., Deroubaix, A. F., Simon, V., German-Retana, S., Legrand, A., Habenstein, B., Zipfel, C., Bayer, E., Mongrand, S. and Germain, V. (2018). REM1.3's phospho-status defines its plasma membrane nanodomain organization and activity in restricting PVX cell-to-cell movement. PLoS Pathog 14(11): e1007378.
- R Core Development Team. (2015). R: A Language and Environment for Statistical Computing. https://doi.org/10.1007/978-3-540-74686-7.
- Sager, R. and Lee, J. Y. (2014). Plasmodesmata in integrated cell signalling: insights from development and environmental signals and stresses. J Exp Bot 65(22): 6337-6358.
- Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., Preibisch, S., Rueden, C., Saalfeld, S., Schmid, B., Tinevez, J. Y., White, D. J., Hartenstein, V., Eliceiri, K., Tomancak, P. and Cardona, A. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods 9(7): 676-682.
- Simpson, C., Thomas, C., Findlay, K., Bayer, E. and Maule, A. J. (2009). An Arabidopsis GPI-anchor plasmodesmal neck protein with callose binding activity and potential to regulate cell-to-cell trafficking. Plant Cell 21(2): 581-594.
- Stahl, Y. and Faulkner, C. (2016). Receptor complex mediated regulation of symplastic traffic. Trends Plant Sci 21(5): 450-459.
- Stahl, Y., Grabowski, S., Bleckmann, A., Kühnemuth, R., Weidtkamp-Peters, S., Pinto, K. G., Kirschner, G. K., Schmid, J. B., Wink, R. H., Hülsewede, A., Felekyan, S., Seidel, C. A. and Simon, R. (2013). Moderation of Arabidopsis root stemness by CLAVATA1 and ARABIDOPSIS CRINKLY4 receptor kinase complexes. Curr Biol 23(5): 362-371.
- Thomas, C. L., Bayer, E. M., Ritzenthaler, C., Fernandez-Calvino, L. and Maule, A. J. (2008). Specific targeting of a plasmodesmal protein affecting cell-to-cell communication. PLoS Biol 6(1): e7.
Article Information
Copyright
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Grison, M. S., Petit, J. D., Glavier, M. and Bayer, E. M. (2020). Quantification of Protein Enrichment at Plasmodesmata. Bio-protocol 10(5): e3545. DOI: 10.21769/BioProtoc.3545.
- Grison, M. S., Kirk, P., Brault, M. L., Wu, X. N., Schulze, W. X., Benitez-Alfonso, Y., Immel, F. and Bayer, E. M. (2019). Plasma membrane-associated receptor-like kinases relocalize to plasmodesmata in response to osmotic stress. Plant Physiol 181(1): 142-160.
Category
Plant Science > Plant cell biology > Intercellular communication
Biochemistry > Protein > Expression
Cell Biology > Cell imaging > Confocal microscopy
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









