- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Cell-free Reconstitution of the Packaging of Cargo Proteins into Vesicles at the trans Golgi Network
(*contributed equally to this work) Published: Vol 10, Iss 5, Mar 5, 2020 DOI: 10.21769/BioProtoc.3537 Views: 5137
Reviewed by: Ralph Thomas BoettcherAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Preparation of Protein Lysates Using Biorthogonal Chemical Reporters for Click Reaction and in-Gel Fluorescence Analysis
Yaxin Xu and Tao Peng
Nov 20, 2024 2011 Views

Sensitive and Adaptable Turn-On Maturation (ATOM) Fluorescent Biosensors for Detecting Subcellular Localization of Protein Targets in Cells
Harsimranjit Sekhon [...] Stewart N. Loh
Mar 20, 2025 2262 Views

Preparation of Testicular Cells for Immunofluorescence Analysis of Manchette in Elongating Spermatids
Changmin Niu [...] Zhibing Zhang
Jun 20, 2025 2374 Views
Abstract
Protein sorting at the trans Golgi network (TGN) plays important roles in targeting newly synthesized proteins to their specific destinations. The aim of this proposal is to reconstitute the packaging of non-Golgi resident cargo proteins into vesicles at the TGN, utilizing rat liver cytosol, semi-intact mammalian cells and nucleotides. The protocol describes how to perform the vesicle formation assay, how to isolate vesicles and how to detect cargo proteins in vesicles. This reconstitution assay can be used to quantitatively measure the efficiency of the packaging of a specific cargo protein into transport vesicles at the TGN under specific experimental conditions.
Keywords: TGNBackground
The trans Golgi network (TGN) is an essential transport hub in the secretory transport pathway. To ensure the fidelity of vesicular trafficking, eukaryotic cells employ a variety of protein sorting machineries to accurately package specific cargo proteins into transport vesicles at the TGN which are then delivered to specific destinations (Guo et al., 2014). To deepen our understanding of the specificity of the TGN sorting process, it is important to develop an assay that can faithfully reconstitute the vesicle formation and cargo sorting process at the TGN. This assay can be utilized to directly and quantitatively measure the roles of a specific factor in regulating the packaging of a specific cargo protein into transport vesicles. Cell-free reconstitution of the packaging of cargo proteins into COPII vesicles from the endoplasmic reticulum (ER) has been well established (Kim et al., 2005; Kim et al., 2007; Merte et al., 2010; Yuan et al., 2018; Niu et al., 2019;). An in vitro assay that reconstitutes the release of a specific cargo protein, TGN46, into transport vesicles at the TGN has been developed (Ponnambalam et al., 1996; Wakana et al., 2012). TGN46 is mainly localized at the TGN, although it can cycle between the TGN and the plasma membrane (Ponnambalam et al., 1996). Recently, we devised an alternative vesicle budding protocol to reconstitute the packaging of non-Golgi resident cargo proteins into vesicles at the TGN (Ma et al., 2018). We performed the vesicle formation assay by incubating cells at 20 °C to accumulate newly synthesized cargo proteins at the TGN and performed the budding reaction in the presence of the GTPase defective mutant, Sar1A(H79G), to inhibit packaging of cargo proteins into Coat Protein Complex II (COPII) vesicles at the ER. Moreover, we utilized floatation to efficiently remove cytosolic proteins that are not associated with vesicles (Figure 1). Using this assay, we have reconstituted release of planar cell polarity proteins, Vangl2 and Frizzled6, from the TGN (Ma et al., 2018). Our assay indicates that the tyrosine sorting motif on Vangl2 and the polybasic sorting motif on Frizzled6 are important for packaging into vesicles (Ma et al., 2018). The GTPase defective mutant form of Arfrp1, Arfrp1 (Q79L), can inhibit the packaging of Vangl2 in vesicles in a concentration dependent manner (Ma et al., 2018).
Materials and Reagents
- Razor blade
- 10 cm cell and tissue culture dishes (Biofil, catalog number: TCD-010100 )
- 15 ml centrifuge tubes (Biofil, catalog number: CFT-011150 )
- Falcon® 50 ml centrifuge tubes (Corning, catalog number: 352070 )
- 1,000 μl tips (USA Scientific, catalog number: 11112021 )
- 200 μl tips (Axygen, catalog number: T200-Y )
- 0.5-10 μl tips (Axygen, catalog number: T300 )
- 1.5 ml microtubes (Axygen, catalog number: 20220415 )
- Axygen® 1.5 ml Maxymum recovery® microcentrifuge tube (low retention) (Corning, Axygen®, catalog number: MCT-150-L-C )
- Polypropylene copolymer ultracentrifuge tube (2.2 ml capacity volume, Hitachi Koki S300536A)
- Polycarbonate tubes, 0.5 ml capacity volume (Beckman Coulter, catalog number: 343776 )
- 200 μl gel loading tips (Thermo Fisher Scientific, catalog number: 010-Q )
- Paragon® Disposable Sterile Blades (Medicom, catalog number: 90010-10 )
- BemisTM ParafilmTM M Laboratory Wrapping Film (Thermo Fisher Scientific, catalog number: 13-374-12 )
- Immobilon®-P transfer membrane PVDF 0.45 μm (Merck, catalog number: IPVH00010 )
- Corning 1 L filter system 0.22 μm (Corning, catalog number: 431098 )
- Sprague-Dawley rats
- Dulbecco's Modified Eagle Medium (DMEM) (Thermo Fisher Scientific, catalog number: 12800082 )
- Opti-MEM (Gibco, catalog number: 31985070 )
- Sodium bicarbonate (Sigma-Aldrich, catalog number: S5761 )
- Fetal bovine serum (FBS) (Thermo Fisher Scientific, catalog number: 10270106 )
- Penicillin streptomycin (Thermo Fisher Scientific, catalog number: 15140122 )
- Polyethylenimine (Polysciences, catalog number: 23966-1 )
- Bio-Rad protein assay dye reagent concentrate (Bio-Rad Laboratories, catalog number: 5000006 )
- Liquid nitrogen
- 0.25% trypsin-EDTA (Thermo Fisher Scientific, catalog number: 25200056 )
- OptiPrepTM density gradient medium (Sigma-Aldrich, catalog number: D1556 )
- Brilliant blue R (Sigma-Aldrich, catalog number: B0149 )
- 2-Mercaptoethanol (βME) (Sigma-Aldrich, catalog number: M6250 )
- 3-color regular range protein marker (Genefist, catalog number: GF6616 )
- Blotting-Grade Blocker (Bio-Rad Laboratories, catalog number: 1706404 )
- Sodium azide (NaN3) (Sigma-Aldrich, catalog number: S8032 )
- SuperSignalTM west pico PLUS chemiluminescent substrate (Thermo Fisher Scientific, catalog number: 34580 )
- HEPES (Acros Organics, catalog number: 172571000 )
- Potassium chloride (KCl) (VWR Chemicals BDH®, catalog number: 2576 )
- Acetic acid, potassium salt (KOAc) (Fisher Scientific, catalog number: 127082 )
- Magnesium acetate tetrahydrate (Mg(OAc)2) (Sigma-Aldrich, catalog number: M0631 )
- D-Sorbitol (Sigma-Aldrich, catalog number: S1876 )
- Sodium dodecyl sulfate (SDS) (Sigma-Aldrich, catalog number: 151213 )
- Bromophenol blue
- Tris (Affymetrix, catalog number: 75825 )
- Glycine
- 40% Acrylamide/Bis Solution (Bio-Rad Laboratories, catalog number: 1610148 )
- Ammonium persulfate (APS) (Sigma-Aldrich, catalog number: A3678 )
- N,N,N′,N′-Tetramethylethylenediamine (Sigma-Aldrich, catalog number: T7024 )
- Sodium dihydrogen phosphate dihydrate (NaH2PO4·2H2O) (VWR Chemicals BDH®, catalog number: 1514L )
- Potassium phosphate dibasic (K2HPO4) (Sigma-Aldrich, catalog number: P3786 )
- Sodium chloride (NaCl) (Sigma-Aldrich, catalog number: S5886 )
- Glycerol (Sigma-Aldrich, catalog number: G7757 )
- Triton® X-100 (Sigma-Aldrich, catalog number: X100 )
- Tween® 20 (Sigma-Aldrich, catalog number: P1379 )
- Protease inhibitor cocktail tablets (Roche, catalog number: 0 5056489001 )
- DL-dithiothreitol (Sigma-Aldrich, catalog number: D9779 )
- Digitonin (Sigma-Aldrich, catalog number: D141 )
- Dimethyl sulfoxide (DMSO) (Sigma-Aldrich, catalog number: D8418 )
- Creatine phosphate (Roche, catalog number: 10621722001 )
- Creatine kinase (Roche, catalog number: 10736988001 )
- Adenosine 5'-triphosphate (ATP) (Sigma-Aldrich, catalog number: 2383 )
- Guanosine 5'-Triphosphate Disodium Salt (GTP) (FUJIFILM Wako Pure, catalog number: 073-03113 )
- Methanol (Scharlab, catalog number: 0 949 )
- Trypsin inhibitor from glycine max (soybean) (Sigma-Aldrich, catalog number: T9003 )
- HyClone® trypan blue solution (GE Healthcare, HyCloneTM, catalog number: SV30084 )
- Antibodies
- Rabbit anti SEC22B was a gift from Prof. Randy Schekman (University of California, Berkeley, CA, USA), and it was used at 1:2,000
- Sheep anti TGN46 (Bio-Rad Laboratories, catalog number: AHP500G ), and it was used at 1:2,000
- Rabbit anti HA (Cell signaling technology, catalog number: 3724 ), and it was used at 1:2,000
- Horseradish peroxidase (HRP)-coupled sheep antibodies against rabbit IgG (GE Healthcare, catalog number: NA934 ), and it was used at 1:5,000
- Anti-sheep IgG–Peroxidase antibody (Sigma-Aldrich, catalog number: A3415 ), and it was used at 1:5,000
- Buffer solutions (see Recipes)
- KHM buffer (1×)
- KHM buffer (2×)
- KHM buffer (10×)
- SDS-PAGE protein loading buffer (5×)
- Transfer buffer
- SDS-PAGE running buffer
- 5% blotting-grade blocker
- PBS
- PBST
- Buffer E
- Lysis buffer
- Stock solutions (see Recipes)
- 10× PBS, pH 7.4
- SDS-PAGE running buffer (10×)
- 1 M KOAc
- 1 M HEPES-KOH, pH 7.2
- 1 M Mg(OAc)2
- 1.5 M Tris-HCl, pH 8.8
- 1 M Tris-HCl, pH 6.8
- 10% SDS
- 10% APS
- 3% NaN3
- 40 mg/ml digitonin
- 100x Protease inhibitors
- 500 mM DTT
- ATP regeneration system (ATP r.s.)
- 10 mM GTP
- 1 mg/ml polyethylenimine
Equipment
- Small beaker
- Dissection scissors
- -80 °C freezer
- Drill (Craftsman 3/8 inch professional electric drill) (Craftsman, model: 315.26946 0 or equivalent)
- Dounce homogenizer (Kimble® 886000-0024 Kontes® 45 ml Potter-Elvehjem Tissue Grinder with PTFE Pestle and Unground Glass Tube, Size: 24, Manufacturer Part No: KIM-886000-0024)
- Eppendorf® 5418R centrifuge, refrigerated, with rotor FA-45-18-11 and rotor lid (Eppendorf®, model: 5418R , catalog number: 5401000013)
- Beckman Coulter high speed centrifuge, with rotor JA-25.50 and rotor lid (Beckman Coulter, model: Avanti® J-E, catalog number: 369001)
- Hitachi ultracentrifuge with T-865 rotor and rotor lid (Hitachi, catalog number: S99978101 )
- Light microscope with a 10× or 20× objective (any simple or compound light microscope is fine)
- Hitachi Koki himac CS150NX micro ultracentrifuge with S55S-2080 rotor, S120A3-2061 rotor and rotor lids (Hitachi, catalog number: HK-CS150NX )
- Barnstead Thermolyne (Thermo Fisher, model: DB16520-26 catalog number: 05852)
- Elite dry bath incubator (Major Science, model: EL-02 catalog number: 95070)
- Eppendorf® 5804R centrifuge with rotor A-4-44 (Eppendorf®, model: 5804R , catalog number: 5805000327)
- Cell disruptor (Disruptor Genie®, catalog number: SI-D268 )
- ChemiDocTM MP Imaging System (Bio-Rad Laboratories, model: ChemiDocTM MP)
Software
- ImageLab software v4.0
- ImageJ
Procedure
Note: All procedures are performed on ice and all centrifugations are performed at 4 °C unless otherwise stated.
- Preparation of cytosol from Rat Liver
- Dissect out livers from a couple of large Sprague-Dawley rats. Each liver typically weighs around 15 g. Usually the final yield per liver is around 15 ml of cytosolic fraction with a protein concentration of 30 mg/ml.
Note: An application to use large Sprague-Dawley rats needs to be submitted and approved by the University of Committee on Use and Care of Animals (UCUCA). - Upon dissection, place each liver in chilled 50 ml Corning tube, rinse three times with cold PBS (around 25 ml per wash).
- Weigh livers, then cut up into a mush with razor blade and dissection scissors on an ice-cold Petri dish. Add 2 ml cold Buffer E per 1 g liver (Recipe A1).
- Homogenize in large Dounce with drill press in batches (around 1 liver per batch) in cold room. 5 strokes with medium-fitting pestle, then 5 strokes with tight-fitting pestle. To be careful, wear goggles. After homogenization, rat livers will be homogenized to tiny pieces and large fragments of rat livers will not be clearly detected.
- Centrifuge the whole homogenate in 50 ml tubes at 1,000 x g for 10 min at 4 °C in Eppendorf® 5418R refrigerated centrifuge using FA-45-18-11 rotor. Collect supernatant and discard pellet.
- Centrifuge the supernatant at 18,459 x g for 20 min at 4 °C in Beckman Coulter high speed centrifuge (Beckman Coulter, Avanti® J-E) using JA-25.50 rotor. Collect supernatant and discard pellet.
- Centrifuge the supernatant at 125,171 x g for 1 h at 4 °C in Hitachi ultracentrifuge (Hitachi ultracentrifuge) using T-865 rotor.
- Take the deep red fraction (avoid pellet and the floating fat layer) and repeat high speed spin 2 or 3 times.
- Determine protein concentration by the Bradford assay and aliquot in 100 μl and 1 ml amounts.
Note: a concentration range of 18-30 mg/ml is considered as a successful extraction. - Snap freeze aliquots in liquid nitrogen and store at -80 °C. Avoid repeated freeze-thaw cycles.
Note: The centrifugation steps to prepare rat liver cytosol are shown in Figure 1.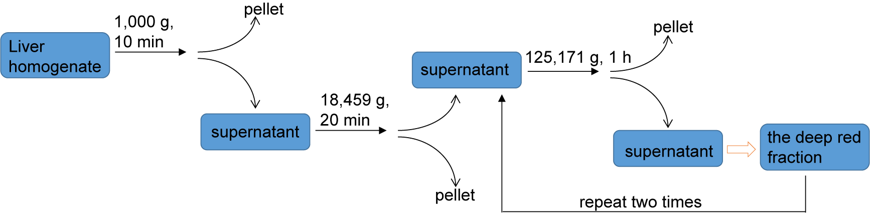
Figure 1. The centrifugation steps to prepare rat liver cytosol
- Dissect out livers from a couple of large Sprague-Dawley rats. Each liver typically weighs around 15 g. Usually the final yield per liver is around 15 ml of cytosolic fraction with a protein concentration of 30 mg/ml.
- Preparation of donor membranes (DM) from cultured HEK 293T cells (prepare on the day of the reaction, Figure 2)
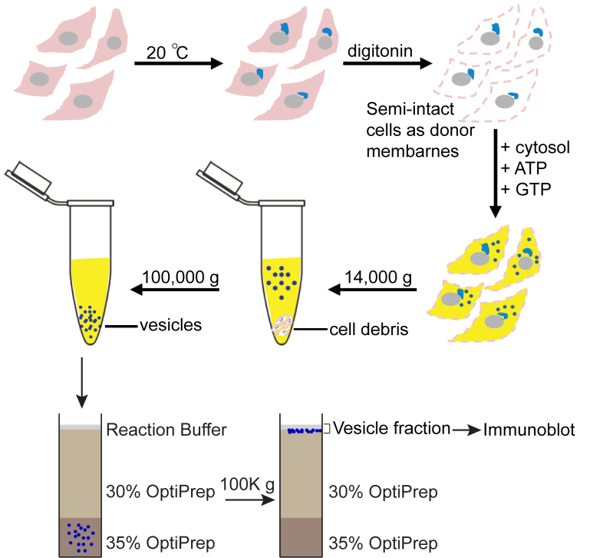
Figure 2. Schematic overview of the assay to reconstitute vesicular release from the TGN. The preparations of semi-intact cells as donor membranes (DM) for the budding reaction and the procedures of the vesicle formation assay and isolation of vesicles produced from semi-intact cells were described in Procedures B and C respectively. Efficiency of the packaging of cargos into vesicles were measured by immunoblotting as described in Procedure D (this Figure is revised from Ma et al., 2018). The DM are also needed for immunoblotting.- Culture two 10 cm plates of HEK 293T cells in GIBCO Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum (FBS), 10 milliunits/ml of penicillin, and 0.1 mg/ml of streptomycin at 37 °C in a cell culture CO2 incubator providing 5% CO2 and around 95% relative humidity. The cells will be grown to 80-100% confluent on the day of the experiment.
Note: Transfect plasmids encoding cargo proteins 24 h before the day of experiment if necessary. - Aspirate media from two 10 cm plates of HEK 293T cells.
- Wash the cells with 5 ml PBS each plate for 2 times.
- Add 10 ml Opti-MEM containing 10% FBS into each plate, seal the plate with parafilm and flow it at 20 °C water bath for 2 h.
- Wash the cells with 5 ml PBS for 2 times for each plate.
- Add 1 ml 0.25% trypsin to each plate and incubate at RT for 2 min.
- Collect cells from each plate with 6 ml PBS buffer into one15 ml Falcon tube.
- Add 25 μl 10 mg/ml trypsin inhibitor to each tube and mix well.
- Centrifuge at 300 x g for 3 min at 4 °C. Aspirate the supernatant and resuspend the cells in 4 ml 1× KHM buffer (Recipe A1) in each tube.
- Add 4 μl digitonin (stock 40 mg/ml in DMSO) to the cells so that final concentration is 40 μg/ml. Mix well and incubate on ice for 5 min.
- Add 8 ml 1× KHM buffer to the cells in each tube, invert the tube and immediately pellet the cells at 300 x g for 3 min at 4 °C.
- Aspirate the supernatant, resuspend the cell pellet in 1 ml 1× KHM buffer in each tube and transfer to a 1.5 ml microcentrifuge tube.
Note: To check if permeabilization is working or not, add 3 μl of cells to 3 μl trypan blue, carefully lay a cover slip over the sample, then check percentage of permeabilized cells under a light microscope with a 16× objective. 100% of cells should be permeabilized at this stage. The nucleus is blue and the intact ER is brown. If the permealization is not working well, a freshly made solution of digitonin is recommended to perform the experiments. - Incubate the cells on ice for 5 min, pellet the cells at 10,000 x g for 5 s at 4 °C and aspirate the supernatant. Resuspend the pellet in 130 μl 1× KHM buffer in each tube and combine the suspensions of semi-intact cells into one 1.5 ml microcentrifuge tube.
- Culture two 10 cm plates of HEK 293T cells in GIBCO Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum (FBS), 10 milliunits/ml of penicillin, and 0.1 mg/ml of streptomycin at 37 °C in a cell culture CO2 incubator providing 5% CO2 and around 95% relative humidity. The cells will be grown to 80-100% confluent on the day of the experiment.
- Reconstitution of the release of vesicles from the TGN
- In low retention tubes, assemble vesicle budding reactions by adding ingredients in Table 1. Each 100 μl reaction contains ATP regeneration system (1 mM ATP, 40 mM creatine phosphate, 0.2 mg/ml creatine phosphokinase in KHM buffer), 0.2 mM GTP, 1 μg Sar1A H79G protein (a GTPase defective mutant version of Sar1A) and rat liver cytosol (4 mg/ml).
Note: Sar1A H79G protein is a C-terminal His-tagged protein purified from E. coli. Add KHM buffer, nucleotides and Sar1A H79G protein first. Mix well by pipetting and briefly centrifuge to collect liquid at the bottom of each tube. Then add semi-intact cells and mix by pipetting up and down gently until homogenous. Add rat liver cytosol last and mix by gentle pipetting.
Table 1. The ingredients added in the TGN vesicle formation assay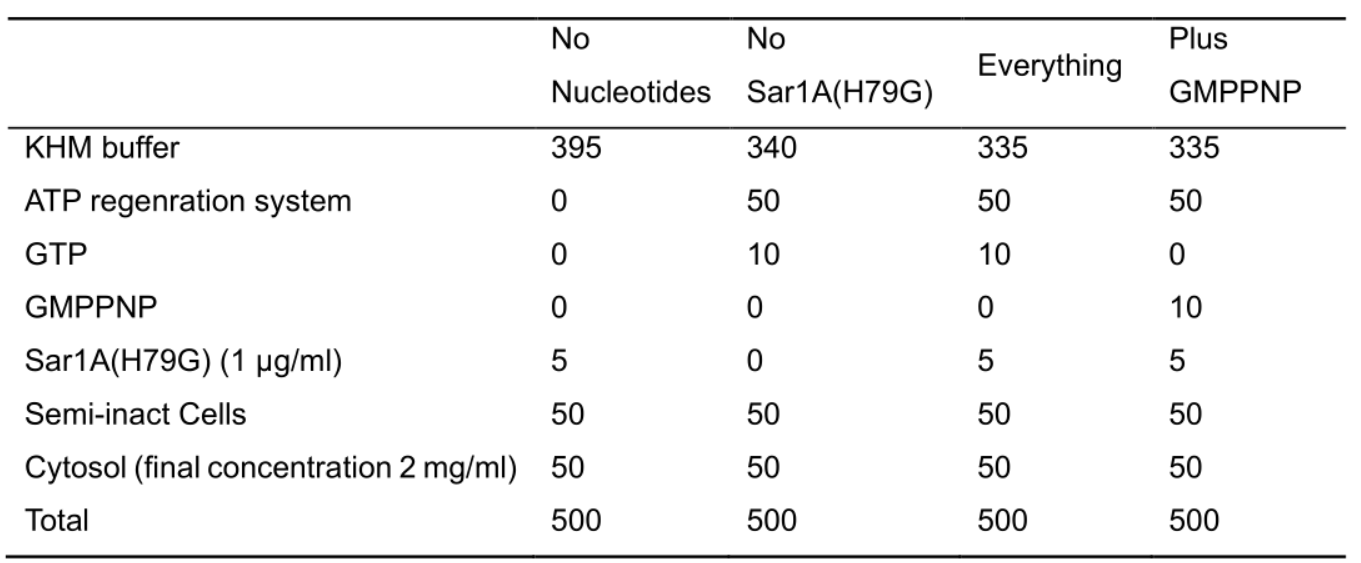
Note: Each column represents a single experimental group. Each row represents a component of the reaction and the volume added to the reaction in μl. The total volume of the reaction is 500 μl. The experimental group without nucleotide is used to monitor the level of cytosolic proteins that are diffused to the vesicle fraction after centrifugation and to monitor the level of cargo proteins that are in the vesicle fraction independent of GTP and ATP. The experimental group performed in the presence of GMPNP, a non-hydrolysable analog of GTP, is to monitor the level of cytosolic and cargo proteins that are present in the vesicle fraction independent of GTP hydrolysis. Sar1A (H79G) is defective in GTP hydrolysis and inhibits packaging of cargo proteins into COPII vesicles. This protein is added to the budding reaction to inhibit COPII-mediated vesicle formation from the ER. - Incubate reactions at 32 °C for 1 h.
- Transfer reactions to ice to stop reaction after incubation.
- Centrifuge at 14,000 x g for 20 min at 4 °C in Eppendorf® 5418R refrigerated centrifuge using FA-45-18-11 rotor to remove cell debris and large membranes.
- Mix supernatant with cold 60% (w/v) OptiPrepTM and 10× KHM buffer (Recipe A3) to reach final concentration at 35% (w/v) OptiPrepTM in 1× KHM buffer.
Note: The OptiPrepTM gradient is a 60% (w/v) solution of iodixanol in water (sterile). - Transfer mixed sample to the bottom of polypropylene copolymer ultracentrifuge tube (2.2 ml capacity volume, Hitachi Koki S300536A) as the bottom layer.
- Overlay with 30% (w/v) OptiPrepTM in 1× KHM buffer [prepared by mixing equal volume of 60% OptiPrepTM and 2× KHM buffer (Recipe A2)] by slowly pipetting against the wall using a gel-loading tip to the ultracentrifuge tube as the medium layer until the tube is almost filled with the whole sample (bottom layer + medium layer).
Note: An interphase should be observed between 30% OptiPrepTM and the supernatant OptiPrepTM mixture at this stage. - Overlay with 50 μl 1× KHM buffer at the top layer by pipetting slowly using a gel-loading tip.
Note: An interphase should be observed between 1× KHM and 30% OptiPrepTM in 1× KHM. - Centrifuge the ultracentrifuge tubes containing the sample at 100,000 x g at 4 °C for 90 min at the Acceleration Setting (Accel) 3 and Decel 4 in Hitachi Koki himac CS150NX micro ultracentrifuge (Hitachi) using S55S-2080 rotor. After centrifugation, the released vesicles will be concentrated at the interface between 1x KHM and 30% OptiPrepTM in 1× KHM.
- Collect vesicles by taking 200 μl supernatant from the top after centrifugation. Transfer the supernatant to thick wall polycarbonate tubes (Beckman Coulter, 0.5 ml capacity volume, 343776 ). Add 300 μl 1x KHM buffer to dilute samples by gently pipetting.
- Centrifuge at 100,000 x g at 4 °C for 30 min in Hitachi Koki himac CS150NX micro ultracentrifuge (Hitachi) using S120AT3-2061 rotor to spin down vesicles.
- In low retention tubes, assemble vesicle budding reactions by adding ingredients in Table 1. Each 100 μl reaction contains ATP regeneration system (1 mM ATP, 40 mM creatine phosphate, 0.2 mg/ml creatine phosphokinase in KHM buffer), 0.2 mM GTP, 1 μg Sar1A H79G protein (a GTPase defective mutant version of Sar1A) and rat liver cytosol (4 mg/ml).
- Immunoblot
Perform standard immunoblotting procedure by the following steps:- Add 20 μl 1× SDS-PAGE protein loading buffer at the bottom of the tube, vortex 8 min to dissolve the vesicles in loading buffer using the cell disruptor (Disruptor Genie®).
- Add DM to 20 μl lysis buffer (Recipe A11) and incubate on ice for 30 min.
- Centrifuge at 14,000 x g for 5 min at 4 °C and collect the supernatant.
- Add 5× SDS-PAGE protein loading buffer (Recipe A4) into the supernatant to reach a final concentration of 1× SDS-PAGE protein loading buffer.
- Incubate both DM sample and high spin pellet (HSP) samples at 55 °C for 30 min.
- Load the DM sample and HSP samples onto a 15 wedged well 10-15% gel according to the cargo protein size.
- Run SDS-PAGE at constant 20 mA until dye runs out of the gel (about 70 min) at RT.
- Transfer protein onto a PVDF membrane at constant 0.3 A for 1-2 h at 4 °C.
- Block the PVDF membrane with 5% blotting-Grade Blocker in PBST (Recipe A7) for 30 min at RT.
- Incubate the PVDF membrane with primary antibodies at RT for 1.5-2 h or at 4 °C overnight.
- Wash with PBST 3 × 5 min.
- Incubate with secondary antibodies conjugated with HRP at RT for 1 h.
- Wash with PBST 3 × 5 min.
- Incubate with the HRP substrate ECL plus and image on a ChemiDocTM Imaging System with ImageLab software v4.0.
Data analysis
- Export immunoblot images from ImageLab software v4.0 as .tif files.
- Use Photoshop and Adobe Illustrator® to process images (Figure 3).
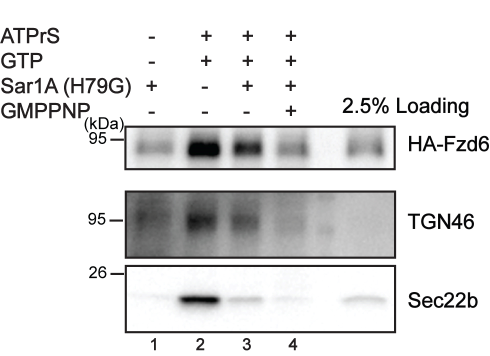
Figure 3. Reconstitution of packaging of Frizzled6 into vesicles from the TGN. COS7 cells were transfected with HA-Frizzled6. On day 1 after transfection, TGN vesicle release reaction was performed using the indicated reagents described in Table 1. The top fraction after flotation was analyzed by immunoblotting with the indicated antibodies. TGN46 and Sec22 are used as positive controls to monitor TGN export and ER export respectively (Republished from Ma et al., 2018).
Recipes
- Buffer solutions
- 1× KHM (1 L)
- Add 5 ml 1 M Mg(OAc)2, 20 ml 1 M HEPES-KOH, pH 7.2 and 110 ml 1 M KOAc to 0.5 L ddH2O
- Adjust volume to 1 L
- Filter the buffer
- Store at 4 °C
- 2× KHM (1 L)
- Add 10 ml 1 M Mg(OAc)2, 40 ml 1 M HEPES-KOH, pH 7.2 and 220 ml 1 M KOAc to 0.5 L ddH2O
- Adjust volume to 1 L
- Filter the buffer
- Store at 4 °C
- 10× KHM (1 L)
- Add 50 ml 1 M Mg(OAc)2 and 200 ml 1 M HEPES-KOH, pH 7.2 to 0.5 L ddH2O
- Dissolve 108 g KOAc
- Adjust volume to 1 L
- Filter the buffer
- Store at 4 °C
- 5× SDS-PAGE protein loading buffer (50 ml)
- Dissolve 3.75 g SDS, 93.75 mg bromophenol blue in a small beaker with 12.5 ml 1 M Tris pH6.8 with constant stirring
- Mix in 25 ml glycerol
- Adjust volume to 37.5 ml with ddH2O
- Aliquot to 750 μl/tube and store at -20 °C
- Add 250 μl βME to an aliquot fresh before use
Note: The final concentration of each reagents in 5× SDS-PAGE protein loading buffer: 7.5% SDS, 50% Glycerol, 250 mM Tris-HCl and 0.19% bromophenol blue, 25% βME (v/v).
- Transfer buffer (2 L)
- Dissolve 6.03 g Tris and 28.8 g glycine in 1.5 L ddH2O (final concentration: 25 mM Tris, 192 mM glycine)
- Add 200 ml Methanol (final concentration: 10%)
- Add ddH2O to 2 L
- Store at 4 °C
- SDS-PAGE running buffer (1 L)
Add 100 ml 10× SDS-PAGE running buffer in 900 ml ddH2O - 5% blotting-grade blocker
Dissolve 5% blotting-grade blocker (w/v) in PBST - PBS (1 L)
Add 100 ml 10× PBS in 900 ml 10× PBS - PBST
0.1% TWEEN® 20 (v/v) in 1× PBS - Buffer E (1 L)
- Add 50 ml 1 M HEPES-KOH, pH 7.2, 70 ml 1 M KOAc and 0.5 ml 1 M Mg(OAc)2 in 800 ml ddH2O
- Dissolve 45.54 g sorbitol and 1.9 g potassium EGTA in the buffer
- Adjust volume to 1 L
- Filter the buffer
- Aliquot to small volume and store at -80 °C
- Add protease inhibitor at 1× concentration and 5 mM DTT before use
- Lysis buffer
Add 0.5% Triton® X-100, 1× protease inhibitor and 1 mM DTT in KHM buffer
- 1× KHM (1 L)
- Stock solutions
- 10× PBS, pH 7.4 (2 L)
- Dissolve 160 g of NaCl, 4 g KCl, 28.8 g Na2HPO4 and 4.8 g KH2PO4 in 1.9 L ddH2O
- Adjust pH to 7.4 with NaOH
- Add ddH2O to 2 L
- Sterilize by autoclaving and store at RT
- 10× SDS-PAGE running buffer (1 L)
- Dissolve 30.3 g Tris, 144.4 g glycine and 10 g SDS in 1 L ddH2O
- Store at RT
- 1 M KOAc (1 L)
- Dissolve 98.14 g potassium acetate 1 L ddH2O
- Sterilize by autoclaving and store at RT
- 1 M HEPES-KOH, pH 7.2 (1 L)
- Dissolve 238.3 g of HEPES in 0.5 L ddH2O
- Adjust pH to 7.2 with KOH
- Add ddH2O to 1 L
- Store at RT
- 1 M Mg(OAc)2 (1 L)
- Dissolve 155.41 g Mg(OAc)2 in 1 L ddH2O
- Sterilize by autoclaving and store at RT
- 1.5 M Tris-HCl, pH 8.8 (1 L)
- Dissolve 181.7 g of Tris in 0.8 L ddH2O
- Adjust pH to 8.8 with HCl
- Add ddH2O to 1 L
- Sterilize by autoclaving and store at RT
- 1 M Tris-HCl, pH 6.8 (1 L)
- Dissolve 121.1 g of Tris in 0.8 L ddH2O
- Adjust pH to 6.8 with HCl
- Add ddH2O to 1 L
- Sterilize by autoclaving and store at RT
- 10% SDS (100 ml)
- Dissolve 10 g of SDS in 90 ml ddH2O
- Store at RT
- 10% APS (50 ml)
- Dissolve 5 g of SDS in 50 ml ddH2O
- Store at 4 °C
- 3% NaN3 (50 ml)
- Dissolve 1.5 g of NaN3 in 50 ml ddH2O
- Store at RT
- 40 mg/ml digitonin (10 ml)
- Dissolve 400 mg digitonin in 10 ml ddH2O
- Aliquot in volumes of 30 μl, store at -20 °C
- 100× protease inhibitors (1 ml)
- Dissolve 2 protease inhibitor cocktail tablet in 1 ml ddH2O
- Aliquot in volumes of 30 μl, store at -20 °C
- 500 mM DTT (1 ml)
- Dissolve 0.077 g DL-dithiothreitol in 1 ml ddH2O
- Aliquot in volumes of 30 μl, store at -20 °C
- ATP regeneration system (20 ml)
- Dissolve 2.04 g creatine phosphate, 40 mg creatine kinase, 101.44 mg ATP in 20 ml 1× KHM
- Aliquot in small volumes and store at -80 °C
- 10 mM GTP (20 ml)
- Dissolve 113.43 mg ATP in 20 ml 1× KHM
- Aliquot in small volumes and store at -80 °C
- 1 mg/ml polyethylenimine (100 ml)
- Dissolve 0.1 g polyethylenimine in 100 ml ddH2O
- Filtered by 0.22 μm filter
- Aliquot in small volumes and store at -80 °C
- 10× PBS, pH 7.4 (2 L)
Acknowledgments
This protocol was modified from previous work described in previous reports (Kim et al., 2005; Yuan et al., 2017; Ma et al., 2018; Niu et al., 2019). This work was supported by the Hong Kong Research Grants Council Grants 26100315, 16102218, 16103319 and 16101116 to Y.G. Y.G. was a previous HHMI postdoctoral research associate during the period of 2009-2013.
Competing interests
The authors declare no conflict of interest or competing interests.
References
- Guo, Y., Sirkis, D. W. and Schekman, R. (2014). Protein sorting at the trans-Golgi network. Annu Rev Cell Dev Biol 30: 169-206.
- Kim, J., Hamamoto, S., Ravazzola, M., Orci, L. and Schekman, R. (2005). Uncoupled packaging of amyloid precursor protein and presenilin 1 into coat protein complex II vesicles. J Biol Chem 280(9): 7758-7768.
- Kim, J., Kleizen, B., Choy, R., Thinakaran, G., Sisodia, S. S. and Schekman, R. W. (2007). Biogenesis of gamma-secretase early in the secretory pathway. J Cell Biol 179(5): 951-963.
- Ma, T., Li, B., Wang, R., Lau, P. K., Huang, Y., Jiang, L., Schekman, R. and Guo, Y. (2018). A mechanism for differential sorting of the planar cell polarity proteins Frizzled6 and Vangl2 at the trans-Golgi network. J Biol Chem 293(22): 8410-8427.
- Merte, J., Jensen, D., Wright, K., Sarsfield, S., Wang, Y., Schekman, R. and Ginty, D. D. (2010). Sec24b selectively sorts Vangl2 to regulate planar cell polarity during neural tube closure. Nat Cell Biol 12(1): 41-46; sup pp 41-48.
- Niu, L., Ma, T., Yang, F., Yan, B., Tang, X., Yin, H., Wu, Q., Huang, Y., Yao, Z. P., Wang, J., Guo, Y. and Hu, J. (2019). Atlastin-mediated membrane tethering is critical for cargo mobility and exit from the endoplasmic reticulum. Proc Natl Acad Sci U S A 116(28): 14029-14038.
- Ponnambalam, S., Girotti, M., Yaspo, M. L., Owen, C. E., Perry, A. C., Suganuma, T., Nilsson, T., Fried, M., Banting, G. and Warren, G. (1996). Primate homologues of rat TGN38: primary structure, expression and functional implications. J Cell Sci 109 (Pt 3): 675-685.
- Wakana, Y., van Galen, J., Meissner, F., Scarpa, M., Polishchuk, R. S., Mann, M. and Malhotra, V. (2012). A new class of carriers that transport selective cargo from the trans Golgi network to the cell surface. EMBO J 31(20): 3976-3990.
- Yuan, L., Baba, S., Bajaj, K. and Schekman, R. (2017). Cell-free generation of copii-coated Procollagen I carriers. Bio-protocol 7(22): e2450.
- Yuan, L., Kenny, S. J., Hemmati, J., Xu, K. and Schekman, R. (2018). TANGO1 and SEC12 are copackaged with procollagen I to facilitate the generation of large COPII carriers. Proc Natl Acad Sci U S A 115(52): E12255-E12264.
Article Information
Copyright
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Tang, X., Yang, F. and Guo, Y. (2020). Cell-free Reconstitution of the Packaging of Cargo Proteins into Vesicles at the trans Golgi Network. Bio-protocol 10(5): e3537. DOI: 10.21769/BioProtoc.3537.
- Ma, T., Li, B., Wang, R., Lau, P. K., Huang, Y., Jiang, L., Schekman, R. and Guo, Y. (2018). A mechanism for differential sorting of the planar cell polarity proteins Frizzled6 and Vangl2 at the trans-Golgi network. J Biol Chem 293(22): 8410-8427.
Category
Molecular Biology > Protein > Detection
Biochemistry > Protein > Posttranslational modification
Cell Biology > Organelle isolation > Golgi
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









