- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Preparation of Single Epithelial Cells Suspension from Mouse Mammary Glands
Published: Vol 10, Iss 4, Feb 20, 2020 DOI: 10.21769/BioProtoc.3530 Views: 6089
Reviewed by: Pooja VermaPia GiovannelliPilar VillacampaAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Thrombopoietin-independent Megakaryocyte Differentiation of Hematopoietic Progenitor Cells from Patients with Myeloproliferative Neoplasms
Chloe A. L. Thompson-Peach [...] Daniel Thomas
Jan 20, 2023 2372 Views

Isolation, Purification, and Culture of Embryonic Melanoblasts from Green Fluorescent Protein–expressing Reporter Mice
Melissa Crawford [...] Lina Dagnino
Sep 5, 2023 2002 Views

Identification and Sorting of Adipose Inflammatory and Metabolically Activated Macrophages in Diet-Induced Obesity
Dan Wu [...] Weidong Wang
Oct 20, 2025 2273 Views
Abstract
Single cell RNA sequencing is a very powerful means for cellular heterogeneous studies and so becoming wildly utilized nowadays. To guarantee the success of such analysis, it is very important, though sometimes difficult, to obtain single cells suspension with high quality, especially from the primary solid organs like mammary glands. Digestion of mouse or human mammary glands with enzymes was previously described. However, the yield, viability, especially the separation degree of the cells have rarely been noticed in these studies. Here we described a detailed protocol for the single epithelial cells suspension preparation from mouse mammary glands, which could be applied for single cell RNA sequencing on different platforms. This protocol could be well adapted for dissociation of other solid organs and tumors, and the single cell suspension could be also used for many other experiments.
Keywords: Single cellsBackground
The mammary gland is very heterogeneous, including many distinct types of cells, such as epithelia, endothelia, adipocytes, fibroblasts and immune cells. The abnormality of epithelia is the main cause for many mammary diseases, like mammary tumors (Prat and Perou, 2009). So, it is of great significance to study the cellular heterogeneity of mammary epithelia in normal and disease conditions. And the single cell RNA sequencing is one of the methods used to study the cellular heterogeneity. The mammary epithelium is mainly composed of two distinct lineages, the basal and luminal cells (Shackleton et al., 2006). According to previous studies, these two cells populations could be well distinguished through flow cytometry analysis for the mouse mammary cells by using cell surface markers CD24 and CD29 (or CD24 and CD49f), as the basal and luminal cells are characterized with CD24MidCD29Hi cells and CD24HiCD29Lo, respectively (Shackleton et al., 2006). Still little attention was paid on the cell numbers, viability and the separation degree of the cells in the flow cytometry analysis and cell sorting. But the cell quality is very important for the single cell RNAseq experiments. To meet this end, we devised a protocol with the combination of the red blood cells’ removal, epithelial cells’ enrichment, and futher cell sorting to separate basal and luminal cells with high cell viability. It’s a better approach to prepare single cells suspension for single cell RNA sequencing. Besides, the protocol has been verified for its adaption towards other solid organs, like liver and brain, as well as tumor/cancer tissues from breast, liver, colon and other organs of mouse and human.
Materials and Reagents
- 50 ml centrifuge tubes (Corning, catalog number: CLS430829)
- 1.5 ml micro-tubes (Axygen, catalog number: MCT150-C-S)
- Pipette tips 10 µl, 200 µl and 1,000 µl (Axygen, catalog numbers: T-400, T-200-Y and T-1000-B)
- Sterile serological pipettes (5 and 10 ml) (Corning, catalog numbers: CLS4487 and CLS4488)
- FACS tubes (FALCON, catalog number: 352058)
- 40 μm cell strainer (FALCON, catalog number: 352340)
- Cell counting chambers (Nexcelom Bioscience, catalog number: CHT4-SD100)
- 0.2 μm Nalgene syringe filter (Thermo Fisher Scientific, catalog number: 725-2520)
- 50 ml syringe (TERUMO, catalog number: SS-50LE)
- Female FVB mice for mammary gland isolation (provided by Animal facility core, Faculty of Health Sciences, University of Macau)
- DMEM/F12 medium (Thermo Fisher Scientific, catalog number: 11330032)
- Fetal Bovine Serum (FBS) (Thermo Fisher Scientific, catalog number: 26140079)
- HBSS (Thermo Fisher Scientific, catalog number: 14140112)
- Hydrocortisone (Sigma-Aldrich, catalog number: H0888)
- Insulin (Sigma-Aldrich, catalog number: 91077C)
- EGF (Thermo Fisher Scientific, catalog number: PHG0313)
- Cholera toxin (Sigma-Aldrich, catalog number: C8052)
- Collagenase Type 3 (Worthington Biochemical Corporation, catalog number: LS004183)
- Hyaluronidase (Sigma-Aldrich, catalog number: H3506)
- Dispase II (Roche, catalog number: 04942078001)
- Deoxyribonuclease I (Worthington Biochemical Corporation, catalog number: LS002145)
- HEPES (1 M) (Thermo Fisher Scientific, catalog number: 15630080)
- DPBS (Thermo Fisher Scientific, catalog number: 14190250)
- 0.25% Trypsin-EDTA (Thermo Fisher Scientific, catalog number: 25200056)
- 1x RBC lysis buffer (Thermo Fisher Scientific, catalog number: 00-4333-57)
- Trypan blue dye (STEMCELL Technologies, catalog number: 07050)
- EasySeq Mouse Epithelial Cell Enrichment kit (STEMCELL Technologies, catalog number: 19868)
- Anti-mouse CD24-PE-Cy7 antibody (BD Biosciences, catalog number:560536)
- Anti-mouse CD29-APC antibody (Biolegend, catalog number:102216)
- DAPI solution (1 mg/ml) (Thermo Fisher Scientific, catalog number: 62248)
- Albumin, Bovine (BSA) (VWR LIFE SCIENCE, catalog number: VWRV0332)
- Digestion buffer A (see Recipes)
- Digestion buffer B (see Recipes)
- FACS buffer (see Recipes)
Equipment
- Sterile scissors and forceps
- Pipette
- Pipette-Aid
- Biosafety cabinet for cell culture work
- Holder for centrifuge tubes
- Humidified cell culture incubator set to 37 °C, 5% CO2
- Orbital shaker (Allsheng, OS-100) settled in cell culture incubator
- Centrifuge with adaptors for 50 ml centrifuge tubes and FACS tubes, for use at room temperature
- EasySep Magnet (STEMCELL Technologies, catalog number: 18000)
- Cell counter (Nexcelom Bioscience, Cellometer Auto 2000)
- BD FACSAria III for cell sorting
Software
- BD FACSDiva 6.1 for BD FACSAria III
Procedure
- Isolation of mammary glands from female mice (Figure 1)
- Sacrifice the female mouse via cervical dislocation and restrain it with abdomen up by pinning its feet into a foam surface using pushpins.
- Make a parallel incision along the mouse’s abdomen without puncturing the peritoneum by using scissors and forceps. Then enlarge the incision along the hind legs to the feet, to make a “Y-type” incision.
- Pull back the skin and pin it to the foam surface to expose the fourth pair of the mammary glands. Remove the whole mammary mounts free from the body.
- Transfer the mammary glands into a sterile 50 ml centrifuge tube and mince it into pieces as small as possible inside the tube with scissors.

Figure 1. Isolation of fourth pairs of mouse mammary glands for cells dissociation. A. The sacrificed mouse was restrained on a foam surface. B. A “Y-type” incision was made to expose the fourth pair of mammary glands (within the red circles). C. The mammary glands were removed and transferred into a 50 ml centrifuge tube. D. The minced mammary tissue was treated with digestion buffer A for cells dissociation.
- Digestion of mammary glands into cell suspension
- Add 5 ml digestion buffer A (for 1 pair of mammary glands) into the 50 ml centrifuge tube with minced mammary pieces. And pipet several times to efficiently mix with a 5 ml sterile serological pipette.
- Put the 50 ml centrifuge tube into the holder of the orbital shaker which is settled in the cell culture incubator (37 °C, 5% CO2). Set the speed as 120 rpm.
- Pipetting the mammary tissue repeatedly (pipetting once every 15-20 min) for sufficient dissociation. It usually takes about one hour.
- Once there are only very few macroscopic pieces left in the tube, spin down the dissociated mammary cells at 300 x g for 3 min at room temperature.
- Remove the supernatant, and re-suspend the cells pellets with 2 ml digestion buffer B. And put the tube on the orbital shaker within the cell culture incubator for 5 min at 120 rpm.
- Spin down the cells and wash them with 5 ml HBSS.
- Spin down again, remove the supernatant and re-suspend the cells pellets with 1 ml 0.25% Trypsin-EDTA for a further digestion on the orbital shaker within the cell culture incubator for 2 min at 120 rpm.
- Neutralize the digestion by adding 9 ml DMEM/F12 medium with 5% FBS and spin down the cells. Wash them once with HBSS and spin down again.
- Epithelial cells enrichment
- To remove the red blood cells, re-suspend the cells pellets with 1 ml 1x RBC lysis buffer and incubate at room temperature for 5 min.
- Stop the reaction by adding 5 ml HBSS. Mix and then spin down the cells.
Note: If the cells pellets appear red or pink color, which indicates there are still some red blood cells left, re-treat the cells with 1 ml 1x Red Blood Cells (RBC) lysis buffer for another 5 min and then wash with HBSS. - Re-suspend the cells pellets with 5 ml DMEM/F12 medium + 5% FBS. Then filter the cells through a 40 µm cell strainer.
- Count the cell number by using the Nexcelom Cellometer Auto 2000 with a cell counting chamber.
- To enrich the mammary epithelial cells, an EasySeq Mouse Epithelial Cell Enrichment kit is used to remove the lineage markers positive (Lin+) non-epithelial cells following the manufacturer’s instructions. Briefly, the Lin+ cells are bound with immune-magnetic beads targeting the lineage markers and attracted to the EasySep magnet, then the desired Lin- epithelial cells remain free for collection.
- Cell sorting to separate basal and luminal cells
- Count the number of the enriched epithelial cells and re-suspend the cells at a concentration of 1 x 106 cells per 100 μl of FACS buffer with anti-mouse CD24-PE-Cy7 antibody (1:200), anti-mouse CD29-APC antibody (1:200) and DAPI (1 μg/ml).
- Stain the cells in ice for 30 min in dark. And then wash cells with DPBS (3-5 volumes of staining buffer) twice (spin down at 300 x g for 3 min).
- Re-suspend the cells with FACS buffer (3 volume as the staining buffer to make a final cell concentration around 3 x 106 cells/ml).
- Load the cells suspension for cell sorting. DAPI-CD24MidCD29Hi cells and DAPI-CD24HiCD29Lo cells were gated and sorted out as basal and luminal mammary epithelial cells, respectively (Shackleton et al., 2006; Sun et al., 2018). The collected basal and luminal cells could be used for single cell RNA sequencing or other purposes.
Data analysis
- A schematic illustration briefly summarizes the major steps of the protocol (Figure 2).
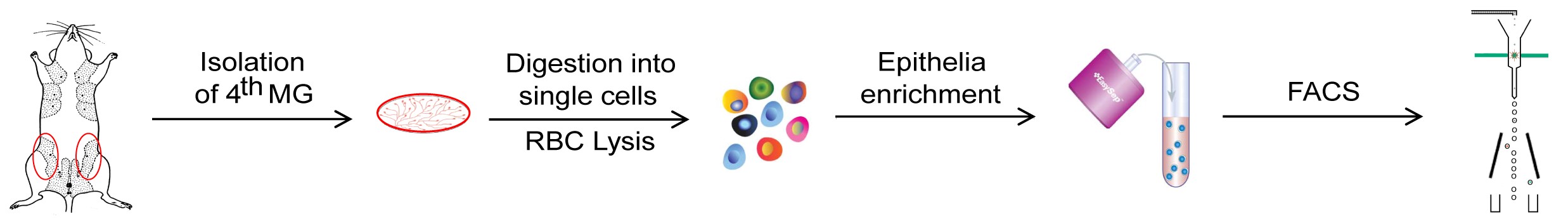
Figure 2. A schematic illustration of the protocol. The fourth pairs of mammary glands were removed from the female mouse and minced into small pieces, then digested into single cells by several enzymes. The red blood cells were lysed and Lin- epithelial cells were enriched. Finally, the basal and luminal mammary cells were separately collected via cell sorting. - The mouse mammary basal and luminal epithelial cells could be well separated via cell sorting based on the distinct expression pattern of cell-surface markers CD24 and CD29 (Sun et al., 2018). As shown in Figure 3, the DAPI-CD24MidCD29Hi cells and DAPI-CD24HiCD29Lo cells were gated as basal and luminal mammary epithelial cells, respectively.

Figure 3. Flow cytometry result of mouse mammary cells. The DAPI- cells were gated out as vital cells (purple color) and then further gated based on distinct expression patterns of CD24-PE-Cy7 and CD29-APC. CD24MidCD29Hi cells were sorted out as basal cells (green color), whereas the CD24HiCD29Lo cells were sorted out as luminal cells (red color).
Notes
- The digestion buffers A and B, and the FACS buffer could be stored in 4 °C for up to 3 months.
- After incised from the body, the mammary gland tissue should be minced and digested as soon as possible to avoid cell viability loss.
- This protocol could be applied in preparation of single cells suspension from other solid organs, like liver and brain, as well as tumor/cancer tissues from breast, liver, colon and other organs of mouse and human.
Recipes
- Digestion buffer A (Table 1)
Table 1. Recipe of Digestion buffer A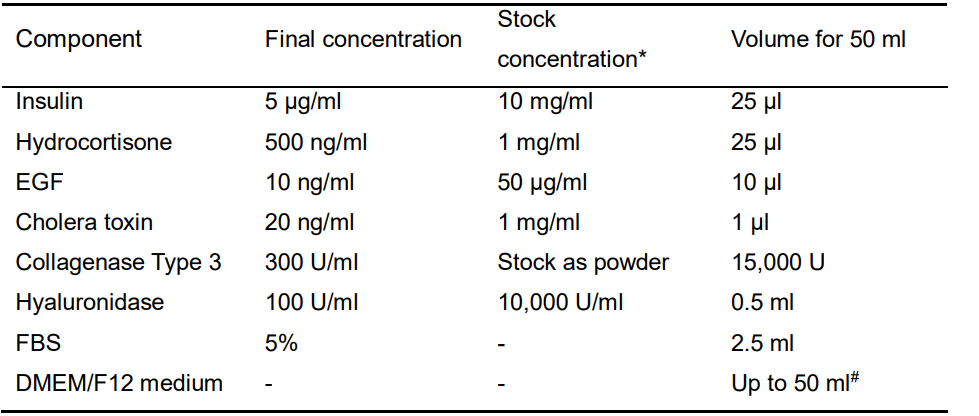
- Digestion buffer B (Table 2)
Table 2. Recipe of Digestion buffer B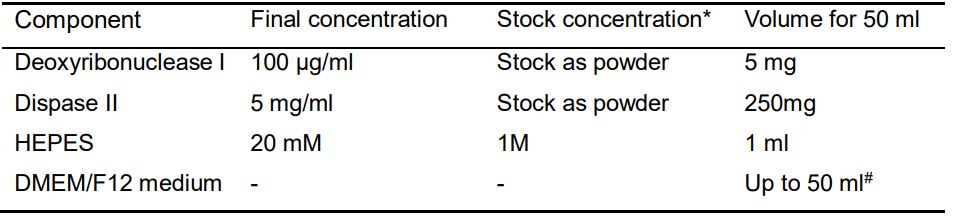
- FACS buffer (Table 3)
Table 3. Recipe of FACS buffer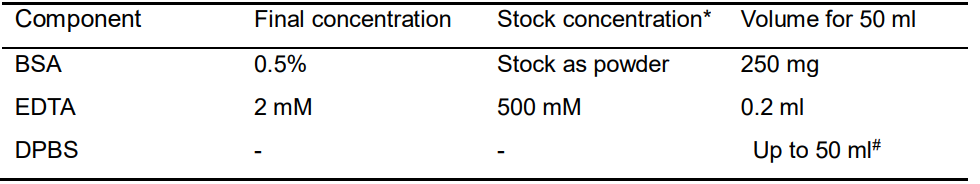
*all the stock reagents should be stored in -80 °C (please use within 1 year), except those as powders, as well as FBS, DMEM/F12 medium and DPBS stored in 4 °C. EDTA could be stored at room temperature.
#all the buffers need to be filtered with 0.2 μm Nalgene syringe filter, and then make the final volume to 50 ml.
Acknowledgments
This protocol was adapted from our previous work (Sun et al., 2018), which was supported by Chair Professor Grant CPG 2017-00016-FHS and Startup Research Grant SRG 2017-00045-FHS, by University of Macau, Macau SAR, China, Macao Science and Technology Development Fund Grants 065/2015/A2 and 094/2015/A3, and by National Natural Science Foundation of China Grant 81602587.
Competing interests
The authors declare that they have no conflict of interest.
Ethics
This protocol involving mice has been approved by the University of Macau Animal Ethics Committee under protocol UMAEC-050-2015 (validate period: 2015-now).
References
- Prat, A. and Perou, C. M. (2009). Mammary development meets cancer genomics. Nat Med 15(8): 842-844.
- Shackleton, M., Vaillant, F., Simpson, K. J., Stingl, J., Smyth, G. K., Asselin-Labat, M. L., Wu, L., Lindeman, G. J. and Visvader, J. E. (2006). Generation of a functional mammary gland from a single stem cell. Nature 439(7072): 84-88.
- Sun, H., Miao, Z., Zhang, X., Chan, U. I., Su, S. M., Guo, S., Wong, C. K. H., Xu, X. and Deng, C. X. (2018). Single-cell RNA-Seq reveals cell heterogeneity and hierarchy within mouse mammary epithelia. J Biol Chem 293(22): 8315-8329.
Article Information
Copyright
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Sun, H., Xu, X. and Deng, C. (2020). Preparation of Single Epithelial Cells Suspension from Mouse Mammary Glands. Bio-protocol 10(4): e3530. DOI: 10.21769/BioProtoc.3530.
- Sun, H., Miao, Z., Zhang, X., Chan, U. I., Su, S. M., Guo, S., Wong, C. K. H., Xu, X. and Deng, C. X. (2018). Single-cell RNA-Seq reveals cell heterogeneity and hierarchy within mouse mammary epithelia. J Biol Chem 293(22): 8315-8329.
Category
Cell Biology > Cell-based analysis > Flow cytometry
Cell Biology > Cell isolation and culture > Cell isolation > Flow cytometry
Cell Biology > Tissue analysis > Tissue isolation
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









