- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Photoactivable Cholesterol as a Tool to Study Interaction of Influenza Virus Hemagglutinin with Cholesterol
Published: Vol 10, Iss 4, Feb 20, 2020 DOI: 10.21769/BioProtoc.3523 Views: 4782
Reviewed by: David PaulAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

A Fluorescence Dequenching-based Liposome Leakage Assay to Measure Membrane Permeabilization by Pore-forming Proteins
Javier Aguilera [...] Jianjun Sun
May 20, 2021 7113 Views

Flotation Assay With Fluorescence Readout to Study Membrane Association of the Enteroviral Peripheral Membrane Protein 2C
Kasturika Shankar [...] Lars-Anders Carlson
Apr 5, 2025 1608 Views

Lipid-Mediated Sequential Recruitment of Proteins Via Dual SLIPT and Dual SLIPTNVOC in Live Cells
Kristina V. Bayer and Richard Wombacher
Nov 5, 2025 1600 Views
Abstract
Non-covalent binding of cholesterol to the transmembrane region of proteins affect their functionalities, but methods to prove such an interaction are rare. We describe our protocol to label the hemagglutinin (HA) of Influenza virus with a cholesterol derivative in living cells or with immunoprecipitated protein. We synthesized a “clickable” photocholesterol compound, which closely mimics authentic cholesterol. It contains a reactive diazirine group that can be activated by UV-illumination to form a covalent bond with amino acids in its vicinity. Incorporation of photocholesterol into HA is then visualized by “clicking” it to a fluorophore, which can be detected in an SDS-gel by fluorescence scanning. This method provides a convenient and practical way to demonstrate cholesterol-binding to other proteins and probably to identify the binding site.
Background
Non-covalent interactions of proteins with cholesterol are supposed to regulate trafficking and functionalities of many proteins (de Vries et al., 2015). However, due to the transient and rather weak nature of this interaction, cholesterol-binding proteins and the respective binding sites are notoriously difficult to identify. Highly sophisticated methods, such as NMR or crystallography require large amounts of purified proteins which need to be integrated into artificial lipid membranes and are thus accessible only to specialized labs. A simple procedure involves measuring the cholesterol content in purified proteins using commercial “kits” (e.g., Amplex red cholesterol assay kit, Molecular Probes). However, this method is insensitive and requires solubilization of proteins from membranes with detergent which often removes non-covalently bound lipids.
One improvement is the synthesis of clickable–photocholesterol compounds which can be covalently linked to a protein. They can be added to cells where they are rapidly integrated into membranes and thus can interact with proteins in their native environment. They contain a diazirine group at position 6 of the sterol ring, which disintegrates upon uv-illumination into molecular nitrogen plus a highly reactive carbene-group that forms a covalent bond with amino acid side chains in close vicinity. To detect cross-linked proteins, probes have a latent affinity handle, an alkyne group for chemical conjugation under physiological conditions to azide-reporters by copper-catalyzed azide-alkyne cycloaddition (“click chemistry”). Reporters are either present on “beads” to enrich and identify probe-interacting proteins by mass spectrometry or are fluorophores which allow their in-gel detection.
We synthesized a compound, 6,6'-Azi-25-ethinylcholesterol [termed improved photoclick-cholesterol, complete synthesis is described in Hu et al. (2019)], which is more similar to genuine cholesterol than a commercially available photoclick-cholesterol (Hex-5'-ynyl 3ß-hydroxy-6-diazirinyl-5α-cholan- 24-oate, Avanti Polar lipids, number 700147) since it contains (besides the alkyne group) no further alterations in cholesterol’s alkyl side chain (Figure 1). A previous study showed that 6-photocholesterol is a faithful mimetic of authentic cholesterol (Mintzer et al., 2002). Note, however, that some of the diazirine groups might be photoactivated to other reactive species that have a longer half time than the carbene-group which might unspecifically label proteins. Thus, photocrosslinking is a qualitative rather than a quantitative measure of the cholesterol affinity of a protein. Nevertheless, both cholesterol probes label only a few specific proteins out of all cellular membrane proteins (Thiele et al., 2000; Hulce et al., 2013), indicating that they are suitable tools for analyzing possible interactions between cholesterol and target proteins.
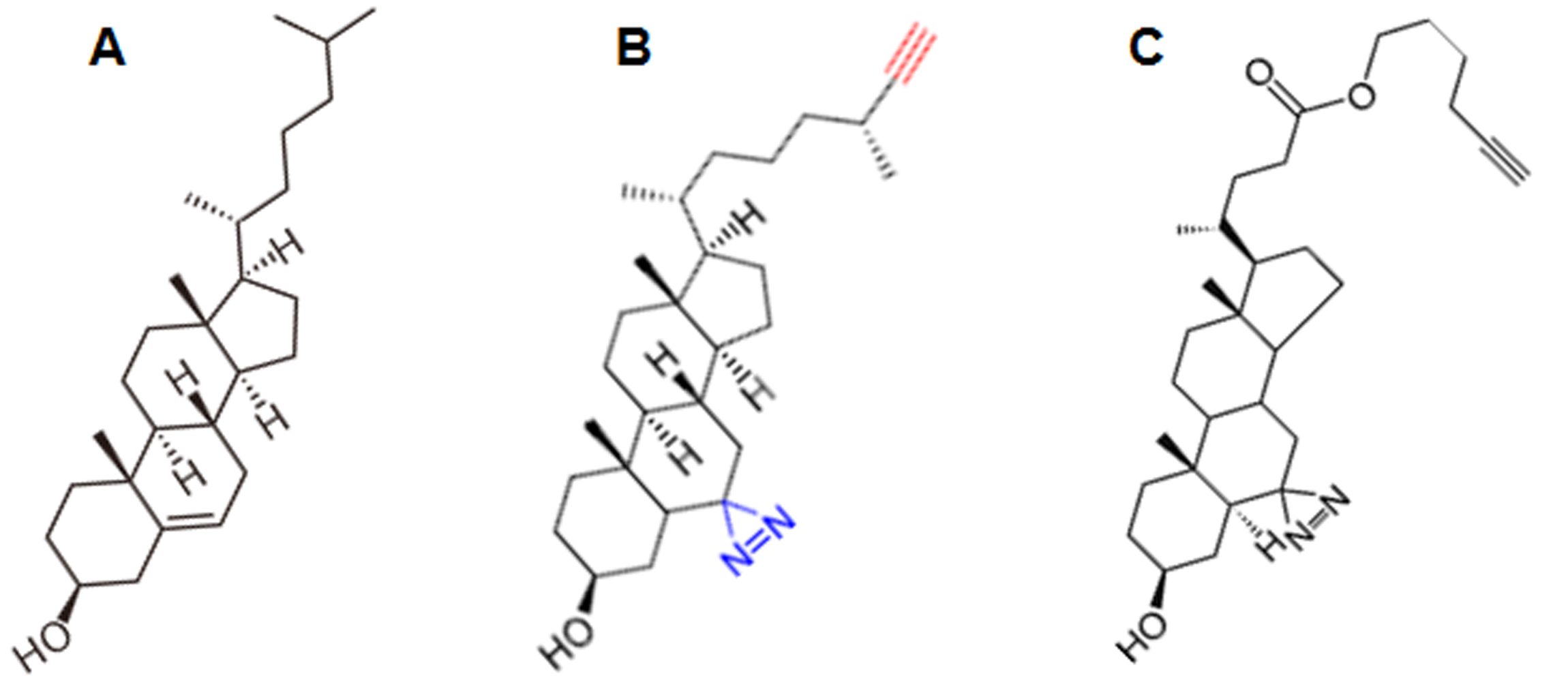
Figure 1. The structure of cholesterol and of two photoactivatable derivatives. A. Cholesterol. B. Improved Photoclick-cholesterol (6,6'-Azi-25-ethinyl-cholesterol) used in this protocol. C. Photoclick-cholesterol (Hex-5'-ynyl 3ß-hydroxy-6-diazirinyl-5α-cholan-24-oate) commercially available from Avanti lipids. The photolabile azide-group is highlighted blue and the alkyne-group in red in B.
Materials and Reagents
- Cell culture consumables
- 6-well/24-well tissue culture plates, flasks, pipettes (Sarstedt, Germany)
- Pipette tips (Sarstedt, Germany)
- Aluminum foil
- CHO-K1 (Chinese hamster ovary cells) (ATCC, catalog number: CCL-61)
- Plasmid (pCAGGS, Niwa et al., 1991)
- DMEM (PAN Biotech, catalog number: P04-04500)
- EDTA-Trypsin (PAN Biotech, catalog number: P10-023100)
- DPBS w/o calcium/magnesium (PAN Biotech, catalog number: P04-36500)
- Penicillin/Streptomycin (10,000 U/ml) (PAN Biotech, catalog number: P06-07100)
- FBS (fetal bovine serum) (PAN Biotech, catalog number: P30-3306)
- Opti-MEMTM I Reduced Serum Medium (Thermo Fisher, GibcoTM, catalog number: 31985070)
- TurboFect Transfection Reagent (Thermo Fisher, Thermo ScientificTM, catalog number: R0531)
- Growth medium (see Recipes)
- Immunoprecipitation
- Ethanol (Sigma, CAS number: 64-17-5, catalog number: 34852-1L-M)
- Tris-HCl (TRIS hydrochloride) (Carl Roth, CAS number: 1185-53-1, catalog number: 9090.3)
- EDTA (AppliChem, CAS number: 6381-92-6, catalog number: A1104)
- Sodium Pyrophosphate (Fisher Scientific, Alfa Aesar, CAS number: 7722-88-5, catalog number: A17546.30)
- Sodium Fluoride (Fisher Scientific, Alfa Aesar, CAS number: 7681-49-4, catalog number: A13019.30)
- Sodium Orthovanadate (Sigma, CAS number: 13721-39-6, catalog number: S6508-10G)
- Benzamidine Hydrochloride (AppliChem, CAS number: 1670-14-0, catalog number: A1380)
- PMSF (AppliChem, CAS number: 329-98-6, catalog number: A0999)
- NEM (N-Ethylmaleimide) (Sigma, CAS number: 128-53-0, catalog number: E3876-5G)
- cOmpleteTM, EDTA-free Protease Inhibitor Cocktail (Sigma, Roche, catalog number: 11873580001)
- NP-40 (Thermo Fisher, Thermo ScientificTM, catalog number: 85124)
- Protein-G-Sepharose 4 Fast Flow (Sigma, GE Healthcare, GE17-0618-01)
- Anti-HA2 antiserum (Hu et al., 2019)
- IP buffer (see Recipes)
- Cell lysis buffer (see Recipes)
- SDS-PAGE and Western blot
- PVDF blotting-membrane, 0.2 µm (VWR, Peqlab, catalog number: 732-3200)
- 1.5 ml microcentrifuge tubes (Sarstedt, Germany)
- 30% acrylamide/ bisacrylamide (37.5:1) (Carl Roth, catalog number: 3029.1)
- Tetramethylethylenediamine (TEMED) (Carl Roth, CAS number: 110-18-9, catalog number: 2367.3)
- Ammonium persulfate (APS) (Carl Roth, CAS number: 7727-54-0, catalog number: 9592.2)
- Sodium dodecyl sulfate (SDS) (Carl Roth, CAS number: 151-21-3, catalog number: 2326.4)
- Bromophenol bule (Sigma, CAS number: 115-39-9, catalog number: 1081220005)
- 1,4-Dithiothreitol (DTT) (Carl Roth, CAS number: 3483-12-3, catalog number: 6908.2)
- Glycerol (Carl Roth, CAS number: 56-81-5, catalog number: 3783.1)
- Skimmed milk powder (Carl Roth, CAS number: 68514-61-4, catalog number: T145.3)
- Tween 20 (Carl Roth, CAS number: 9005-64-5, catalog number: 9127.1)
- PierceTM ECL Plus Western Blotting Substrate (Thermo Fisher, catalog number: 32132)
- Stacking-gel solution (see Recipes)
- Separating-gel solution (see Recipes)
- 5x Non-reducing loading buffer (see Recipes)
- 4x Reducing loading buffer (see Recipes)
- Blocking buffer (see Recipes)
- Crosslinking and click chemistry
- Blacklight Blue (UV) Lamp (Sankyo Denki, power 8W, 3.5 A, 60V, wavelength 320-365 nm, Figure 3A)
- Improved Photoclick-cholesterol (Figure 1B, dissolved in ethanol, the tube is sealed and stored at -20 °C). The long synthesis is described in all details in Hu et al. (2019). But an intermediate alkyne-cholesterol can be purchased from Click Chemistry Tools (https://clickchemistrytools.com/product/alkyne-cholesterol/). With this compound it might be easier to synthesize the final product, the improved Photoclick-cholesterol.
- Picolyl-Azide-Sulfo-Cy3 (Jena Bioscience, catalog number: CLK-1178-1)
- CuAAC Biomolecule Reaction Buffer Kit (THPTA-based) (Jena Bioscience, catalog number: CLK-1178-1)
Equipment
- -20 °C freezer
- Pipette controller (Hirschmann Laborgeräte)
- Heracell 240i CO2 incubator (Heraeus)
- Tabletop centrifuge (Eppendorf, model: 5424R)
- Power pack p25 (Analytik Jena, Biometra P25T)
- Chemiluminescence Imaging, Fusion SL (PeqLab)
- PerfectBlueTM Semi-Dry Electro Blotting Systems (VWR, PeqLab, model: SedecTM M)
- Tube-rotator (Carl Roth, ThermoFisher, model: ATX1.1)
- Typhoon FLA 9400 scanner (GE Healthcare) (Figure 2)

Figure 2. Typhoon FLA 9400 scanner. The gel from Part I Section D, which contains the cross-linked samples and washed with ddH2O, is inserted between two glass plates for scanning with a laser. For results, see Figures 4A and 4B.
Software
- Typhoon Scanner Control v5.0 Software (GE Healthcare)
- ImageQuant (GE Healthcare)
- ImageJ (Fiji)
- Microsoft Excel
- GraphPad Prism
Procedure
Part I: Photocholesterol labeling of HA in living cells
- HA expression in CHO cells by transfection
- Seed 0.9 x 106-1.1 x 106 CHO cells into one well of a 6-well plate one day before transfection such that they reach 70-90% confluency the next day.
- Dilute 3 µg plasmid (pCAGGS) encoding HA in 300 µl serum-free Opti-MEM and mix by pipetting.
- Briefly vortex TurboFect reagent and add 6 µl to the diluted DNA. Mix immediately by vortexing.
- Incubate 15-20 min at room temperature.
- During the incubation time, remove the old cell medium and replace by 1 ml serum-free DMEM.
- Add the DNA/TurboFect reagent mixture dropwise to the cell medium.
- Gently rock the plate to achieve an even distribution of the DNA/TurboFect complexes.
- Incubate at 37 °C in a CO2 incubator.
- Photocholesterol labeling of HA inside cells and crosslinking
- At 6 h post transfection, replace the cell medium with 1 ml fresh serum-free DMEM to remove the transfection reagent.
- Add 5 μl photocholesterol (from a 5 mg/ml stock in ethanol, which is stored at -20 °C, final concentration: 50 μM) to each well and gently rock the plate to achieve an even distribution.
- Incubate at 37 °C in a CO2 incubator until the next step.
- At 24 h post transfection, replace the cell medium with 1 ml fresh serum-free DMEM to remove dead cells floating in the culture supernatant.
- Put the plate in the cold room (~8 °C, Figure 3B) or on ice as long as the “UV lamp/6-well plate/ice box” can be protected from light. Cold temperature slows down the mobility of lipids within the membrane and prevents conformational changes of proteins, and is thus better suited for subsequent crosslinking.
- Take off the lid from the plate and from the UV lamp since plastic will absorb UV-light (Figure 3C).
- Put the UV lamp directly on top of the 6-well plate, turn on the UV lamp (wavelength 320-365 nm, power 8 W, 3.5 A, 60 V) to expose cells to UV light for 10-20 min in the dark to activate the diazirine group to trigger crosslinking (Figure 3D). The cells are still attached to the 6-well culture plate and are covered by 1 ml serum-free DMEM at this stage. Wear protective googles during illumination.
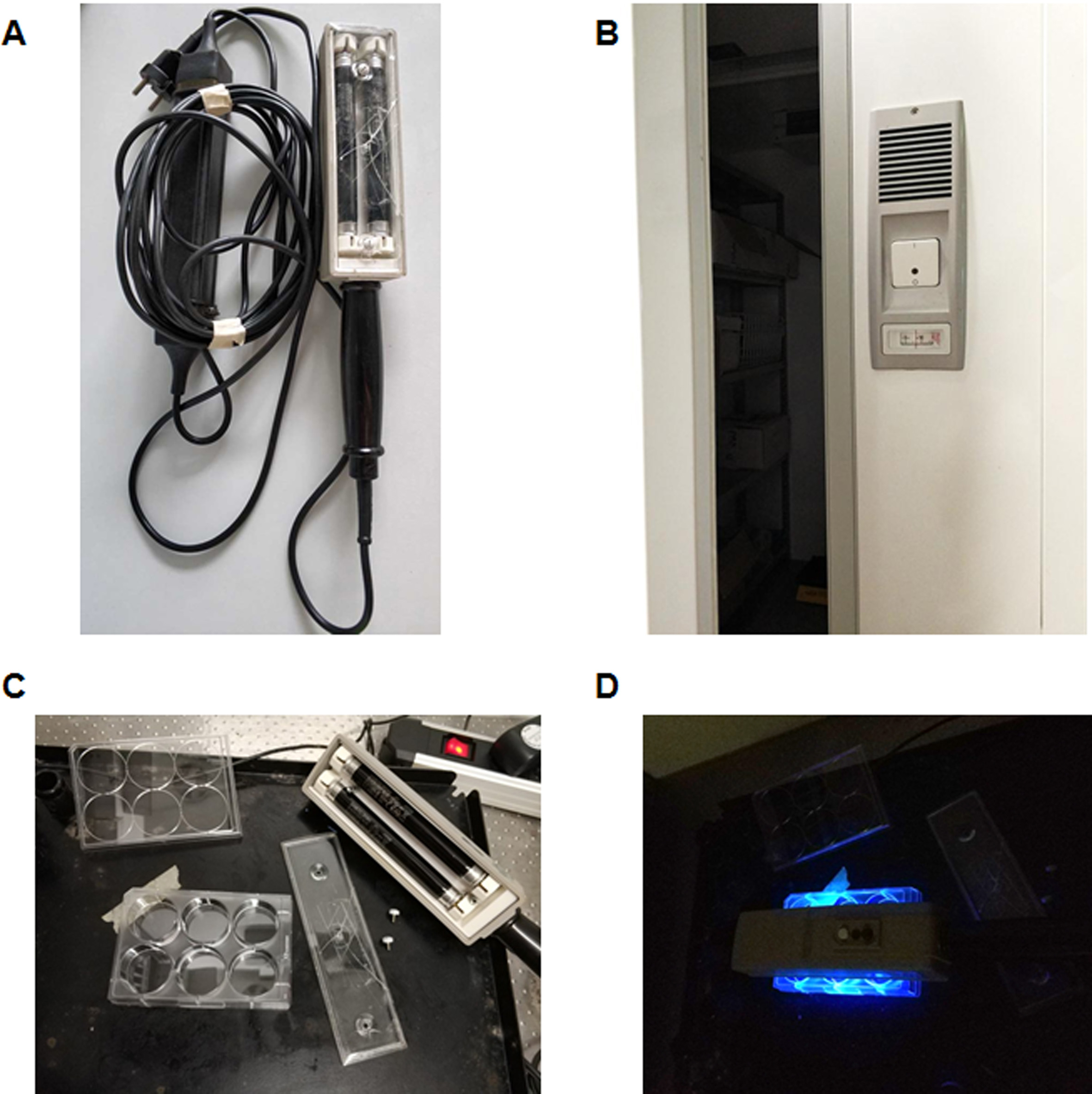
Figure 3. Crosslinking with photocholesterol. A. UV lamp (wavelength 320-365 nm, power 8 W, 3.5 A, 60 V). B. Cold room (Tm: ~8 °C). C. Preparation before UV illumination by taking off the lid from the tissue culture plate and from the UV lamp. D. UV illumination of the culture plates in the dark.
- Immunoprecipitation of HA (buffers should be cold and procedures are performed at 4 °C)
- After exposure to UV light cells are still attached to the 6-well culture plate and are covered in DMEM. Put the 6-well plate on ice and detach the cells by scraping without changing medium.
- Transfer cells to 1.5 ml microcentrifuge tubes and centrifuge at 1,200 x g for 5 min at 4 °C.
- Wash the cell pellet once with 1 ml PBS and resuspend the cells in 500 μl cell lysis buffer.
- Lyse the cells for 30 min at 4 °C and centrifuge at maximal speed (13,500 rpm/17,000 x g ) for 15 min at 4 °C to remove insoluble material.
- Transfer the supernatant to a new 1.5 ml microcentrifuge tube without disturbing the pellet.
- Remove 50 μl from the lysate for analysis of protein expression by Western blot. Add 12 µl 5x non-reducing loading buffer to this 50 μl lysate, incubate at 95 °C for 5 min and store at -20 °C.
- Add anti-HA2 antiserum (1:1,000) to the remaining 450 μl lysate and incubate at 4 °C with agitation overnight (12-16 h) in the cold room.
- Prepare working solution of protein-G-sepharose beads according to the manufacturer’s instruction.
a.The stock of protein-G-sepharose is stored in ethanol. Thus, centrifuge the stock solution at 12,000 x g for 30 s to remove the ethanol.
b.Wash the protein-G-sepharose beads once with PBS by adding the same volume of PBS to the beads and centrifuge at 12,000 x g for 30 s.
c.Resuspend the protein-G-sepharose beads in PBS by adding the same volume of PBS (1:1 dilution). - Add 50 μl of the protein-G-sepharose beads to each sample and incubate at 4 °C for another 4 h in the cold room with agitation.
- Centrifuge at 12,000 x g for 30 s to pellet the HA/antibody/protein-G-sepharose complex.
- Remove the supernatant and wash the complex twice with 1 ml IP-buffer and then twice with 1 ml PBS to exclude the possibility that chemicals from the IP-buffer affect the click chemistry reaction in the next step (described in Part I Section D). For the same reason, remove the liquid as completely as possible after the last washing with PBS.
- Proceed to the click chemistry reaction (described in Part I Section D) as soon as possible. Storing the HA/antibody/protein-G-sepharose complex on ice for 1 h is fine, but storing for longer time (e.g., overnight at -20 °C) is not recommended.
- (Cu(I))-catalyzed Azide-Alkyne Click chemistry reactions (CuAAC) according to the manufacturer’s instruction
- Prepare stock solutions in ddH2O: 250 mM THPTA, 100 mM CuSO4, 1 M Na-Ascorbate. The efficiency of the reaction is higher when all the reagents are freshly prepared.
- Prepare stock solution of Picolyl-Azide-Sulfo-Cy3 (2 mM, dissolved in H2O, red color).
- Prepare CuSO4:THPTA-premix freshly (molar ratio of 1:5) for each experiment. 6 µl (2 µl CuSO4 + 4 µl THPTA) mixture is used here for each reaction. (The ratio of CuSO4:THPTA is recommended as 1:5 in the manufacturer’s instruction, but could be changed.)
- Resuspend HA/antibody/protein-G-sepharose complex in 164 µl of H2O.
- Add 10 µl of 2 mM Picolyl-Azide-Sulfo-Cy3 to the mixture and mix by pipetting.
Note: The amount of Azide-Cy3 added here is adjusted based on the amount used in labeling of immunoprecipitated HA. The same final concentration of photocholesterol (50 µM) is used for HA labeling. The molar ratio of alkyne:azide in click chemistry reaction is 1:4, which could be adjusted to get a better result. A higher ratio would give a stronger fluorescence signal. A ratio of 1:10 is recommended in the manufacturer’s instruction. - Add 6 µl CuSO4:THPTA-premix to the mixture and mix by pipetting.
- Add 20 µl Na-Ascorbate to the mixture, such that the final volume of each reaction is 200 µl. Mix by pipetting.
- Incubate samples at room temperature for 1 h with gentle mixing (avoid vortexing) every 10-15 min. Protect samples from light by wrapping the tubes in aluminum foil.
- Wash samples with 1 ml PBS (at least 4 times) until no red color from Azide-Cy3 is seen in the supernatant. However, extensive washing of the beads is not recommended since the amount of beads becomes slightly reduced after each washing step.
- Resuspend samples in 40 µl 1x reducing loading buffer (diluted from 4x reducing loading buffer in ddH2O) and incubate at 95 °C for 5 min.
- Run SDS-PAGE.
- Wash the gel with ddH2O for at least 1 h with periodic changes (e.g., 10 min each). Sometimes overnight (> 12 h) washing is required to reduce the background signal from uncoupled Picolyl-Azide-Sulfo-Cy3.
- Scan the gel with a biomolecular imager (Typhoon FLA 9400 scanner, Excitation = 550 nm, Emission = 560 nm for Cy3).
Note: The order of Steps D4-D7 should not be changed.
Part II: Photocholesterol labeling of immunoprecipitated HA
Transfect CHO cells to express HA as described in Section A of Part I. 24 h after transfection, immunoprecipitate HA with anti-HA2 antiserum as described in Section C of Part I, and then proceed to crosslinking.
Crosslinking between immunoprecipitated HA and photocholesterol:
- Resuspend the washed HA/antibody/protein-A-sepharose complex in 100 µl PBS.
- Add 0.5 µl photocholesterol (from a 5 mg/ml stock dissolved in ethanol, final concentration: 50 µM), and mix by pipetting. Transfer the solution into one well of a 24-well plate such that a thin liquid layer covering the bottom of the well is formed.
- Expose samples to UV light (see Figure 3).
- Transfer the labeled protein to a microcentrifuge tube and wash the mixture twice with 1 ml IP buffer and then twice with 1 ml PBS.
- Proceed to click chemistry reaction as described in Section D of Part I.
Part III: Western blot to determine the expression level of HA
- Prepare gel for SDS-PAGE. The concentration of acrylamide in the stacking gel is 4%, while that in the separation gel is 12%.
- Run SDS-PAGE at constant voltage. Use low voltage (80-100 V) while proteins are in the stacking gel and high voltage (160 V) for the separation gel.
- Transfer the proteins from the SDS-gel onto a polyvinylidene difluoride (PVDF) membrane at 200-220 mA for 1 h using a semi-dry transfer system.
Note: The time could be adjusted based on the molecular weight of the protein under study. Larger proteins require longer transfer time. - Incubate the membrane with blocking solution for 30 min at room temperature.
- Incubate the membrane with primary antibody (anti-HA2 antiserum, diluted 1:1,000 in blocking solution) for 2 h at room temperature or overnight (12-16 h) at 4 °C.
- Wash the membrane 4 times (5 min each) with PBST (see Recipes) on an orbital shaker.
- Incubate the membrane with secondary antibody coupled to horseradish peroxidase (1:5,000 dilution in blocking solution) for 1 h at room temperature.
- Repeat Step 6 in Part III.
- Apply the ECL plus reagent to the membrane according to manufacturer’s instruction, which reacts with horseradish peroxidase to give the chemiluminescent signals.
- Detect the protein with a Fusion SL camera system.
Data analysis
- For quantification of band intensities, Western blot and the fluorogram were analyzed with ImageJ (Fiji) software. The ratios of photo-cross-linking (fluorogram) to protein expression (Western blot) were calculated for a HA-mutant lacking the cholesterol binding site, and results were normalized to values of HA wild type (HA-wt, set to 100%).
- For detailed instruction of using ImageJ please go to https://imagej.net/ImageJ. Here we briefly describe the process of quantification (see Figure 4C). 1) Make sure that the bands are horizontally aligned. 2) Subtract the background signal from cells transfected with empty vector. 3) Select the bands [the left panel in Figure 4C(b) and second panel in Figure 4C(c)]. 4) Plot the band, which gives histograms indicating the intensity of each bands [the middle panel in Figure 4C(b) and the third panel in Figure 4C(c)]. 5) Draw lines to seal each histogram. 6) Select the “magic wand” button and click inside each histogram, which will give a value indicating the intensity of the analyzed band in a new “Result” window [the last panel in Figure 4C(b) and Figure 4C(c)]. Values are exported as csv. file and analyzed by Microsoft Excel (see Table 1). Cholesterol incorporation into the HA-mutant lacking the cholesterol-binding site is reduced to 34% relative to HA-wt.
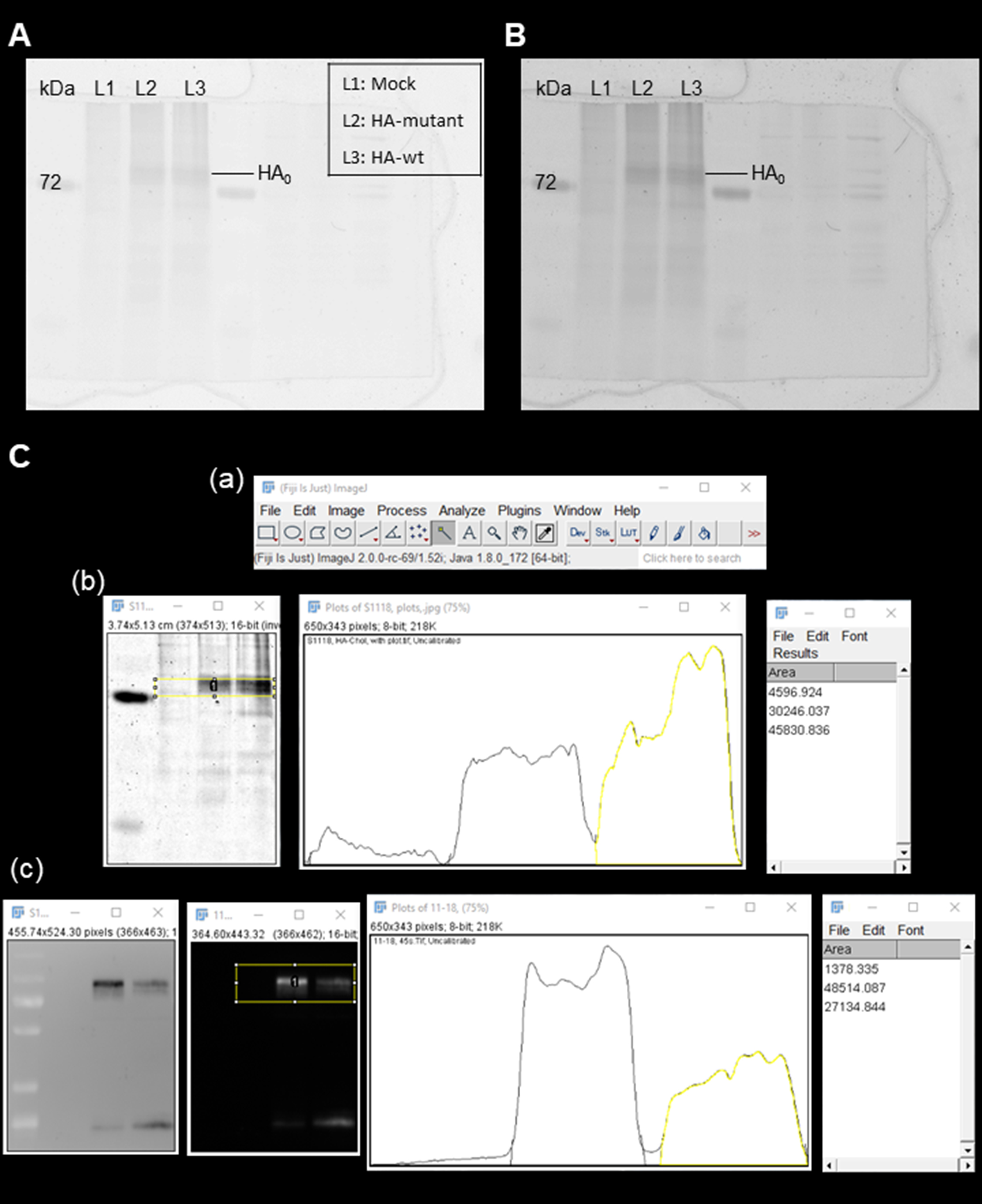
Figure 4. Image processing. A-B. Increasing the voltage from 600 (A) to 800 (B) of the scanner increases the contrast. The molecular weight is shown on the left. The next three lanes (L1, L2 and L3) are samples from cells transfected with empty vector (mock), plasmid expressing a HA mutant lacking the cholesterol binding site (HA-mutant), and plasmid expressing the wild-type HA (HA-wt), respectively. The scan can be exported as a tif-file and evaluated with ImageJ. C. Images processed by ImageJ (Fiji). (a) The working panel of ImageJ (Fiji). (b-c) Quantification of the band intensity of HA-mutant and HA-wt from fluorescence scanner (b) and Western blot (c). The density of bands in the yellow boxes is determined and the area under the curves is calculated for each peak. The results obtained from cells transfected with empty vector (mock) is subtracted and the relative incorporation of photo-cholesterol into the HA-mutant relative to HA-wt is calculated (Table 1).
Table 1. Data processing to compare photocholesterol incorporation into HA-mutant with HA-wt (an example)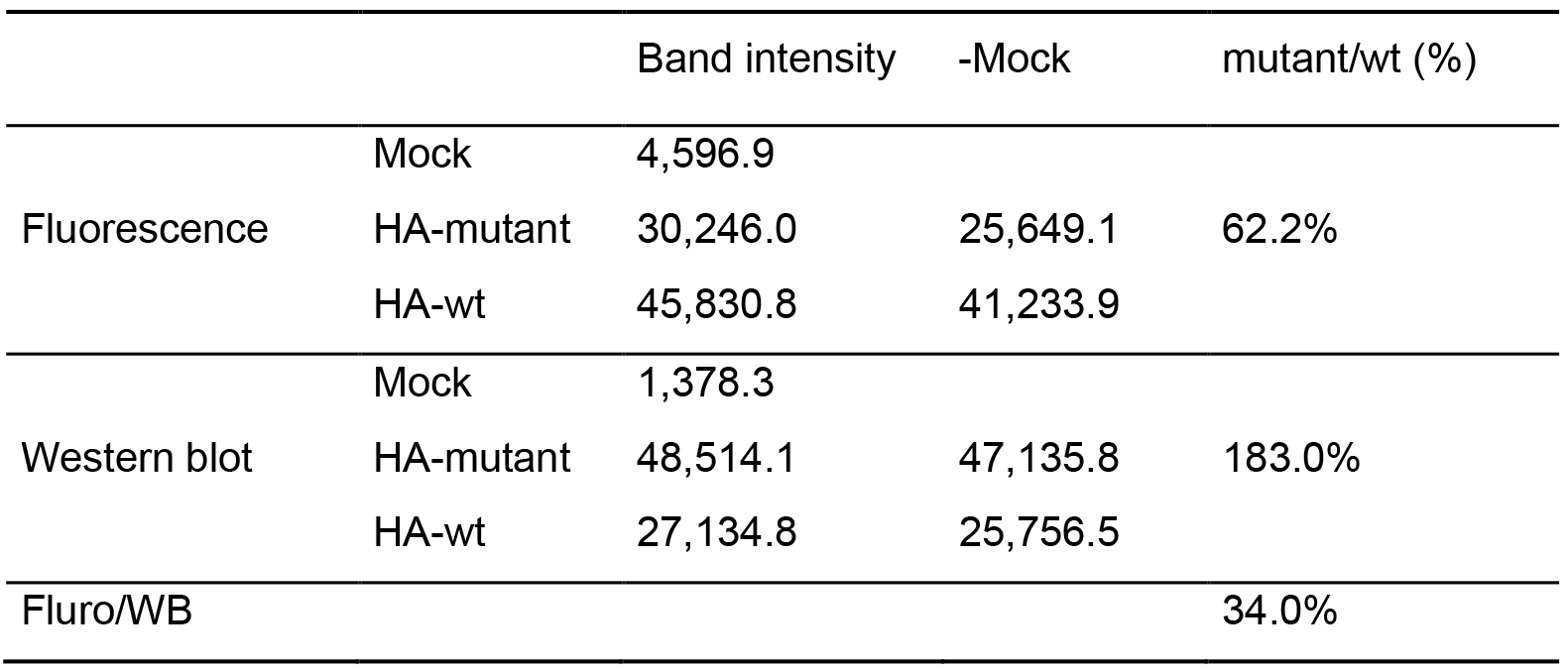
Notes
- Since the cholesterol concentration in membranes is very high (up to 50% in the plasma membrane) and since both lipids and proteins are mobile within the membrane, every transmembrane region is likely to have transient contact with cholesterol. Thus, in principle every transmembrane protein can be crosslinked to photocholesterol, especially if it is strongly expressed. In order to exclude such an “unspecific” interaction, it is advisable to include a mutant in which the supposed cholesterol-binding site is mutated. In the case of HA labeling, a mutant with exchange of the site was reduced to 58%, relative to wt HA and normalized for the expression level of both proteins.
- Since HA with mutations in the cholesterol-binding site are transported to the plasma membrane, but are no longer present in cholesterol-enriched domains (de Vries et al., 2015), one can argue that its reduced labeling inside cells is due to the fact that less photocholesterol is present in their immediate neighborhood. Therefore, we also labeled immunoprecipitated HA, which, however, also revealed a reduced incorporation of photocholesterol (38% relative to wt HA).
Trouble shooting:
- Efficient cross-linking requires large amount of the target protein. Therefore, the corresponding gene should be cloned into a vector with a strong promoter to achieve a high expression level.
- The amount of photocholesterol used for labeling can be varied. We tested 20 µM, 50 µM and 100 µM (final concentration) for labeling with immunoprecipitated HA. Higher concentration gives stronger fluorescence signal in the gel. In the final protocol, we used the same final concentration of 50 µM for labeling HA both in living cells (5 µl photocholesterol in 1 ml DMEM) and with immunoprecipitated protein (0.5 µl photocholesterol in 0.1 ml PBS).
- If no signal is seen in the target protein after labeling of living cells, one could still try labeling of the immunoprecipitated protein since higher concentration of photcholesterol can be achieved and the compound does not compete with authentic cholesterol present in the membrane.
- If the analyzed protein is unstable, the step of washing with PBS (Step D9 in Part I) can be omitted. 4x reducing loading buffer can be directly added to the samples, which are boiled and subjected to SDS-PAGE. In this case, overnight washing of the gel with ddH2O is necessary to wash away background signal from Picolyl-Azide-Sulfo-Cy3.
- If a positive result cannot be repeated, consider to use a new CuAAC kit.
Recipes
- Growth medium (stored at 4 °C)
DMEM
10% FCS
100 U/L Penicillin/Streptomycin - IP buffer (filtered and stored at 4 °C for up to 1 year)
500 mM Tris-HCl (pH 7.4)
20 mM EDTA
30 mM sodium pyrophosphate decahydrate
10 mM sodium fluoride
1 mM sodium orthovanadate
2 mM benzamidine
1 mM PMSF (phenylmethylsulfonyl fluoride)
1 mM NEM
1x protease inhibitor cocktail (freshly added for each experiment)
Note: Some protease inhibitor could be omitted depending on the studied protein. - Lysis buffer (freshly prepared)
1% NP40 in IP buffer - Stacking-gel solution (freshly prepared)
4% (w/v) acrylamide/bisacrylamide
0.1% SDS
125 mM Tris-HCl (pH 6.8)
0.075% (w/v) APS
0.15% (v/v) TEMED - Separating-gel solution (freshly prepared)
12% (w/v) acrylamide/bisacrylamide stock solution
0.1% (w/v) SDS
375 mM Tris-HCl (pH 8.8)
0.05% (w/v) APS
0.1% (v/v) TEMED - 5x non-reducing loading buffer (stored at -20 °C)
1 M Tris-HCl
10% (w/v) SDS
50% (v/v) glycerin
0.1 % (w/v) bromophenol blue
pH 6.8 - 4x reducing loading buffer (stored at -20 °C)
1 ml 5x non-reducing buffer
250 µl 2 M (20x) DTT - PBST (stored at room temperature)
PBS with 0.1% Tween-20 - Blocking solution (freshly prepared)
3% skimmed milk in PBST
Acknowledgments
This work was supported by the German Research Foundation (SFB 740, TP C3) and by the Human Frontiers Science Program (awarded to M.V.). B.H. is the recipient of a Ph.D. fellowship from the China Scholarship Council (CSC). C.T. was funded by the German Research Foundation under Germany’s Excellence Strategy (EXC2151-390873048). M.R.G is recipient of a Ph.D. fellowship from the DAAD. This protocol was adapted and modified from Hu et al. (2019).
Competing interests
B.H. and co-authors declare no conflicts of interest.
References
- de Vries, M., Herrmann, A. and Veit, M. (2015). A cholesterol consensus motif is required for efficient intracellular transport and raft association of a group 2 HA from influenza virus. Biochem J 465(2): 305-314.
- Hu, B., Höfer, C. T., Thiele, C. and Veit, M. (2019). Cholesterol Binding to the Transmembrane Region of a Group 2 Hemagglutinin (HA) of Influenza Virus Is Essential for Virus Replication, Affecting both Virus Assembly and HA Fusion Activity. J Virol 93(15): e00555-00519.
- Hulce, J. J., Cognetta, A. B., Niphakis, M. J., Tully, S. E. and Cravatt, B. F. (2013). Proteome-wide mapping of cholesterol-interacting proteins in mammalian cells. Nat Methods 10(3): 259-264.
- Mintzer, E. A., Waarts, B. L., Wilschut, J. and Bittman, R. (2002). Behavior of a photoactivatable analog of cholesterol, 6-photocholesterol, in model membranes. FEBS Lett 510(3): 181-184.
- Niwa, H., Yamamura, K. and Miyazaki, J. (1991). Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108(2): 193-199.
- Thiele, C., Hannah, M. J., Fahrenholz, F. and Huttner, W. B. (2000). Cholesterol binds to synaptophysin and is required for biogenesis of synaptic vesicles. Nat Cell Biol 2(1): 42-49.
Article Information
Copyright
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Hu, B., Gadalla, M. R., Thiele, C. and Veit, M. (2020). Photoactivable Cholesterol as a Tool to Study Interaction of Influenza Virus Hemagglutinin with Cholesterol. Bio-protocol 10(4): e3523. DOI: 10.21769/BioProtoc.3523.
Category
Biochemistry > Lipid > Lipid-virus interaction
Biochemistry > Protein > Interaction > Protein-lipid interaction
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








