- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Surface Plasmon Resonance Analysis of the Protein-protein Binding Specificity Using Autolab ESPIRIT
Published: Vol 10, Iss 4, Feb 20, 2020 DOI: 10.21769/BioProtoc.3519 Views: 8722
Reviewed by: Alka MehraAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

MicroScale Thermophoresis as a Tool to Study Protein-peptide Interactions in the Context of Large Eukaryotic Protein Complexes
Maximilian G. Plach [...] Thomas Schubert
Dec 5, 2017 24625 Views

CRISPR-mediated Tagging with BirA Allows Proximity Labeling in Toxoplasma gondii
Shaojun Long [...] L. David Sibley
Mar 20, 2018 12491 Views

Isolation of Antigen-Specific Nanobodies From Synthetic Libraries Using a Protein Selection Strategy That Combines MACS-Based Screening of YSD and FLI-TRAP
Apisitt Thaiprayoon [...] Dujduan Waraho-Zhmayev
Jan 20, 2026 452 Views
Abstract
Direct protein-protein interactions are known to regulate a wide range of cellular activities. To understand these contacts one can employ various experimental methods like Dynamic Light Scattering (DLS), Fluorescence Resonance Energy Transfer (FRET), Isothermal titration calorimetry (ITC), Chemical crosslinking, Co-immunoprecipitation (Co-IP), Surface Plasmon Resonance (SPR) and many more. Among these, SPR stands out as a quick, label-free, reliable, and accurate quantitation technique. We have used SPR to elucidate the linkage between 14-3-3 Protein 3 (EhP3) and the actin cytoskeleton in the protist pathogen Entamoeba histolytica. It allowed us to screen EhP3 binding with several actin-binding/actin regulatory proteins (Coactosin, Actophorin, Twinfilin, Profilin, and Filamin). Our screening results suggested Coactosin as an important interacting partner of EhP3. A complete kinetic analysis indeed confirmed that EhCoactosin binds EhP3 with an affinity constant of 3 μM.
Keywords: SPRBackground
Surface Plasmon Resonance (SPR) technique has emerged as one of the most promising screening tools to study macromolecular interactions, since its inception in the early 1990s (Nguyen et al., 2015). It helps determine binding affinities, kinetic parameters, and specificity of these interactions in real-time. This technique is simply based on the optical property of light and principally measures the change in the refractive index upon binding of any molecule to the surface. Surface Plasmon Resonance utilizes the interaction occurring between light and matter. The SPR signal is determined at the surface of the sensor, thus it can be easily correlated to the macromolecules bound on it. This technique captures the real time binding between interacting molecules in a label-free manner, thus making it easy to perform and analyze. Moreover, unlike ITC, this technique is not solution based and uses solid surface for ligand immobilization. SPR uses low quantities of reagents, thus working with small sample size is not an issue.
Principle: Surface Plasmon is a plane-polarized electromagnetic wave that travels on the surface at the interface of the metal coating of the sensor disk (gold disk) and the dielectric medium (sample layer/buffer). These waves are created by continuous fluctuation of charge at the metallic surface. The thin metal surface is placed incident to a laser beam. When the incident light strikes at the interface of the two dielectric media (gold layer and sample layer interface), total internal reflection occurs. Simultaneously, this light generates an evanescent field, with its maximum intensity at the surface of the dielectric material. The resonance occurs when the free electrons of the metal oscillate (plasmon wave) and absorb the plane-polarized light at the specific angle (SPR angle). At the SPR angle, a sharp decrease in the intensity of the reflected light (since some of light is transferred to the Plasmon wave) is observed and this serves as the measurement key. Light passes through a prism and reaches the surface (gold surface) where the molecules are bound. Prism is required to make sure that the wave vector of light in air (kx) is same as the wave vector at the noble metal surface (ksp). The wave vector parallel to the surface is of sole importance in an SPR experiment since the plasmons are confined to the plane of the gold surface. The relationship between kx and incidence angle can be stated as:
kx = k0 nglass sinθinc
where k0 can be calculated as k0 = 2πλ0-1, nglass is the refractive index of the glass prism and θinc is the angle of incidence.
To make the laser light hit the gold disk surface directly, immersion oil is used between the disk and the hemi-cylinder prism (Figure 1). The refractive index of the prism glass, immersion oil, and the disk are same i.e. 1.5. The sensing surface (gold surface) has the lowest refractive index, thus the position of the SPR angle depends on it. Any change (for example: protein-protein interactions typically have a refractive index increment of about 0.18-0.19 ml/g) (Davis et al., 2000) in the dielectric constant or the refractive index changes the resonance angle thus making the SPR-effect a useful tool for us. As the macromolecules bind the surface, the refractive index changes, changing the SPR angle. This change is directly sensed by the detector and translated into response units. The response units obtained are then used to analyze the binding kinetics. The shift in the SPR angle shares a linear relationship with the amount of sample bound. 120 m° (millidegrees) change represents a change in surface protein mass of ~1 ng/mm2. Since light does not penetrate the sample, any colored, turbid, or opaque sample can be used to study.
Figure 1. Pictorial representation of the Surface Plasmon Resonance principle. The diagram depicts the basic underlying principle of the plasmon wave on the gold surface. The Amine groups and the Thiol groups represent the two surface activation method available. The smiley represents the immobilized ligand protein. The chemical structures are labeled to depict the surface chemistry. ksp represents the surface wave vector and kx is the x component of the light wave vector. Absorbed light represents the light at angle of minimum reflection that is recorded as the absorbance by the detector. Evanescent field is a comparable electric field generated by the plasmons on either side of the surface. SPR angle and the Angle of reflection can be clearly seen as read by the detector.
Instrument: Figure 2A shows the Autolab ESPRIT instrument used for the SPR studies. The machine is simple to handle with easy mechanism of operation. The hemi-cylinder is the prism from where the incident light enters and strikes the gold disk. The cuvette (marked as Channel 1 and 2 in the Figure 2B) above it is where all the solutions dispensed in the 384-well plate are injected over the gold disk for the interaction studies to be measured. The syringe pumps modulate the movement of these solutions in and out of the two needles. The Auto sampler moves the needles to and fro for the same. Channel 1 is experimental whereas Channel 2 serves as the control. Simultaneously, the peristaltic pumps below the rack holding the 384-well plate pump in and out the Running buffer. Figure 2C shows the hemi-cylinder with the gold disk placed on it. The Gold Sensor Disk used here is glass disk coated with a very thin layer of gold. The SPR effect can be observed only in the metals where its electrons can behave like a free electron gas. This means that when these electrons move over the surface, their movement is not dependent on the charge that they leave behind. Thus, the only option of metallic surfaces is restricted to Copper, Aluminium, Silver, and Gold.
Figure 2. Autolab ESPRIT instrument. A. The instrument used in the protocol is labeled here. B. Enlarged view of the cuvette, where the reagents are fed onto the gold sensor disk. C. This figure is used from the Autolab manual. The hemi-cylinder is mounted with the gold disks, as placed inside the machine.
Another instrument, commonly used for SPR studies is Biacore T200, sold by G.E. Healthcare. The difference lies at the mechanism of measurement of the resonance angle. Biacore uses a convergent beam, thereby generating a number of incidence and reflecting angles. The disadvantage of this mechanism is that it decreases the resolution to 10-3 in the refractive index. Autolab ESPRIT on the other hand the incident light can be changed by the use of vibrating mirror system. The critical angle for total internal reflection can be changed using the mirrors and the angular shift is measured for a non-coated gold sensor surface with a resolution of approximately 0.02 millidegrees (m°), corresponding to a refractive index resolution of approximately 1 x 10-5. For a coated gold sensor surface, the angular shift is measured with a resolution of approximately 0.1 millidegrees (m°); corresponds to 2 x 10-6 refractive index resolution.
The Chemical steps
Baseline: The first step is to measure the response units for the coupling buffer on the gold sensor disk and is done by simple passing the coupling buffer over the gold surface. It helps in normalization of the further measurements to subtract the noise from the buffer.
Activation: In this step (Figure 3) the gold surface is chemically activated so that protein can be conjugated to it. It can be done by several ways, the most commonly used for example, are: Amine coupling: With the use of EDC (Dimethylaminopropyl-N′-Ethylcarbodiimide N-3-hydrochloride) and NHS (N-Hydroxy Succinimide), the surface can be activated followed by attachment of the protein through its NH2 group. The final concentrations used are 400 mM and 100 mM for EDC and NHS respectively. Thiol coupling: If the ligand protein has a thiol group, then the surface can be made active by using PDEA (2-(2-pyr-idinyldithio) ethaneamine hydrochloride)).
Coupling: At this step, the protein is injected and allowed to bind to the activated surface. This step is termed as immobilization of the ligand protein. As the ligand protein binds, the SPR angle changes linearly with increase in concentration and is measured in its response units.
Blocking: A blocking compound, essentially Ethanolamine here, is added to coat any remaining activated site on the sensor to prevent non-specific binding during the SPR reaction. The blocked sites will be as shown in Figure 1.
Regeneration: This is primarily washing off of any unbound chemical and to set the new baseline value after protein immobilization.
Figure 3. Gold surface activation reactions for the sensor disks. A. The steps for the amine coupling of the ligand protein to the gold surface. B. The steps for the thiol coupling of the ligand protein, using respective chemical reagents. The smiley represents the ligand protein to be immobilized.
Once these steps are successfully performed, the analyte is then passed over the ligand protein to perform the SPR assay. The following Figure 4. represents a typical layout of the reaction and its steps. Here, the running buffer (Association Buffer) is used to set the baseline value since the analyte is prepared in this buffer. As the analyte is injected, the response units increase upon its binding to the ligand protein. Once it reaches a steady state, the dissociation kinetics is measured by just flowing the running buffer over the sensor chip, allowing the analyte to dissociate from the bound ligand. Finally, the regeneration buffer is used to remove the bound analyte to free the ligand protein for the next set of binding reactions.
Figure 4. Steps in a typical SPR assay. Running buffer serves as the baseline for the entire reaction. The Response Units (RU) measured for the running buffer is used to normalize all the calculations. If the injected analyte binds to the ligand protein, then the increase in RU is recorded, while the dissociation of the same analyte leads to the decreasing RU. Regeneration buffer removes any bound analyte (if any) and prepares the ligand for the next round of association reaction.
Relative to other established methods for monitoring protein-protein interactions; SPR-based techniques allow easy experimental designs, rapid testing of hypotheses with high sensitivity, and accurate quantification. In a recent study published by us, SPR was used to determine the interacting partners for EhP3, in order to decipher its connection with the actin cytoskeletal system in Entamoeba histolytica. We conclusively found that EhCoactosin binds EhP3 with an affinity constant of 3 μM (Kumar et al., 2014; Agarwal et al., 2019). This protocol is a one-step handout for anyone who wishes to perform SPR using the Autolab ESPRIT machine. We have explained all the steps in an elaborate manner to help the user to carryout a hassle free experiment. The mechanisms of this instrument are different from Biacore T200 and this protocol can be easily followed by, even a first time user.
Materials and Reagents
- Bare gold sensor disk (Metrohm Autolab, catalog number: 1-04-04-000)
- 384-well Clear Flat Bottom Polystyrene Not Treated Microplate, with Lid, Sterile (Corning, catalog number: 3680)
- 6-well Clear Not Treated Multiple Well Plates (Corning Costar, catalog number: 3736)
- Immersion oil
- Demineralized (demi) water (Sigma-Aldrich, catalog number: 38796)
- Mercaptoundecanoic acid (11-MUA) (Sigma-Aldrich, catalog number: 450561, MW 218.36 g/Mol)
- N-Hydroxy Succinimide (NHS) (Sigma-Aldrich, catalog number: 130672, MW 115.09 g/Mol)
- Dimethylaminopropyl-N′Ethylcarbodiimide N-3-hydrochloride (EDC) (Sigma-Aldrich, catalog number: 03450, MW 191.70 g/Mol)
- Ethanolamine (Sigma-Aldrich, catalog number: E6133, MW 97.54 g/Mol)
- Sodium acetate trihydrate (Sigma-Aldrich, catalog number: S8625, MW 136.08 g/Mol)
- Acetic acid (Sigma-Aldrich, catalog number: A6283)
- HEPES sodium salt (Sigma-Aldrich, catalog number: H7006, MW 260.29 g/Mol)
- Ethanol, ≥ 99.9% (Merck, catalog number: 100983)
- Sodium chloride (NaCl) (Sigma-Aldrich, catalog number: S7653, MW 58.44 g/Mol)
- Sodium hydroxide (NaOH) (Merck, catalog number: 1.06498, MW 40.00 g/Mol)
- Hydrochloric acid (HCl), 37% (Merck, catalog number: 100317)
- Tween 20 (Sigma-Aldrich, catalog number: P1379)
- Nitrogen gas (Sigma-Aldrich, catalog number: 295574)
- 11-MUA solution (see Recipes)
- 400 mM EDC (see Recipes)
- 100 mM NHS (see Recipes)
- 1 M Ethanolamine (see Recipes)
- 1 M Acetate buffer (see Recipes)
- 1 M HEPES buffer (see Recipes)
- 5 M NaCl (see Recipes)
- 1 M NaOH (see Recipes)
- 10% Tween 20 (see Recipes)
- Buffer 1: Coupling buffer (see Recipes)
- Buffer 2: Association buffer (see Recipes)
- Buffer 3: Regeneration buffer (see Recipes)
- Protein solutions (see Recipes)
Equipment
- Tweezers
- Pipettes
- Autolab SPR ESPRIT (Metrohm, model: Autolab ESPRIT)
- Tabletop high-speed centrifuge (Eppendorf, model: 5430R)
- Filtration unit (Milipore, model: XX1514700, and simple Syringe filters)
Software
- Autolab Data Acquisition program
- Autolab Kinetic Evaluation program
Procedure
- Preparation of the gold disk sensor (Figure 5)
- Place the bare gold disk carefully in a 6-well plate using a pair of tweezers, keeping the gold surface facing upwards.
- Add the solution of 11-MUA (Recipe 1) and cover the plate carefully.
- Incubate the disk overnight in this solution to obtain a uniform layer of thiols over the gold surface. The plate is covered with foil and incubated at room temperature.
- Next day, wash three times with ethanol to remove excess thiol groups. For this, pipette out the 11-MUA solution and pipette in ethanol. The volume of the ethanol should be just enough to completely immerse the disk. Gently shake the plate to rinse the disk.
- To remove ethanol, wash thrice with demineralized water, while gently shaking the plate for rinsing the gold disk.
- After thorough washing, blow-dry the disk by flushing the well with nitrogen gas.
- These disks can now be stored up to 2 months in their container at room temperature.
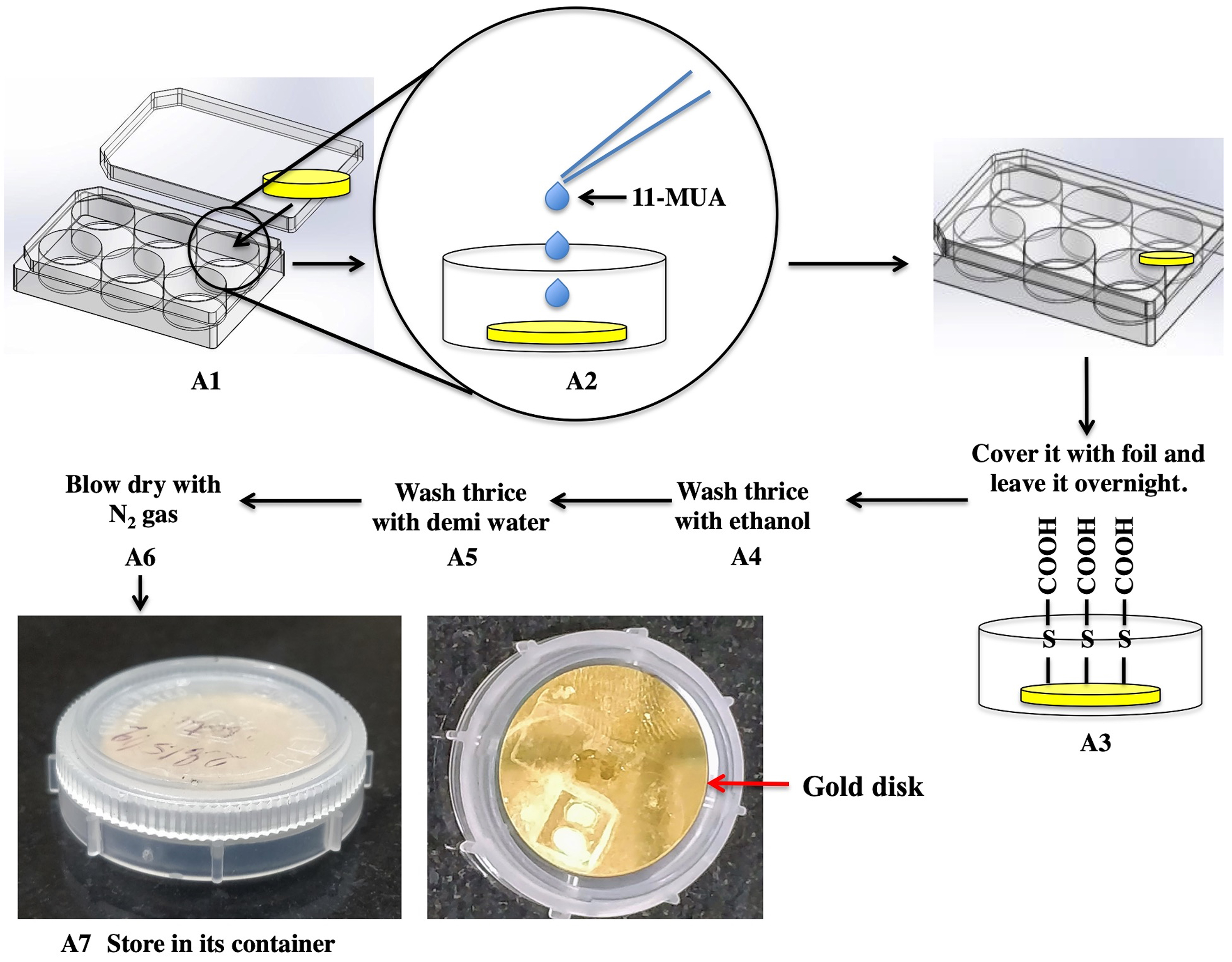
Figure 5. Preparation of the gold sensor disk. The diagram explains all the steps for the Steps A1 to A7 as mentioned in the text.
- Setting up the sensor disk (Figure 6)
- Put a small drop of the immersion oil on the outer edge of the hemi-cylinder.
- Place the 11-MUA coated gold disk on the other end using a pair of tweezers. Now, simply push the disk carefully by sliding till it is correctly set on the hemi-cylinder.
- Make sure the gold surface is facing upwards.
- No air bubble should be present between the disk and the immersion oil on the hemi-cylinder.
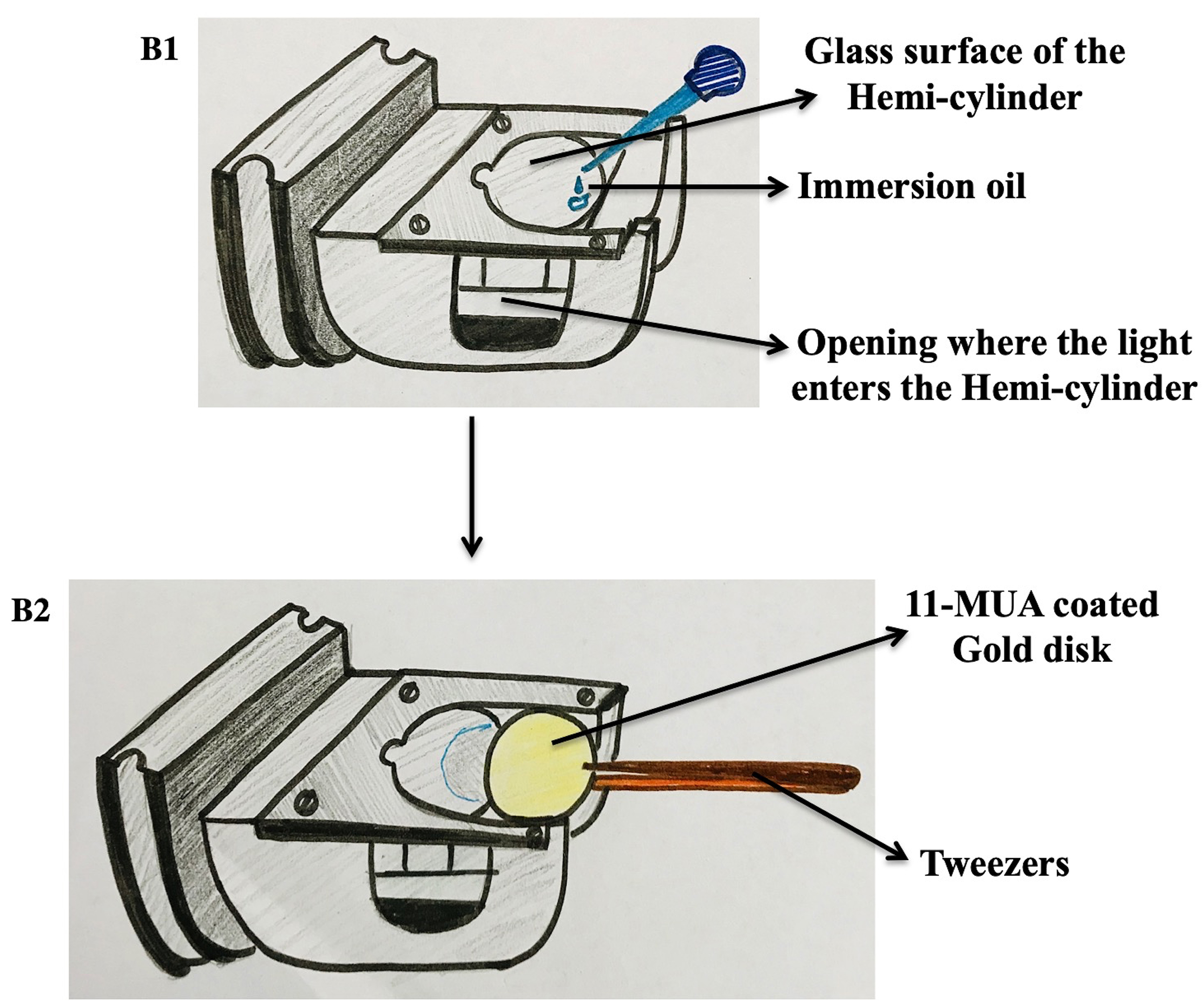
Figure 6. Setting up the gold disk on the hemi-cylinder. B1. The hemi-cylinder is prepared by adding a drop of immersion oil on it. B2. The coated gold disk is placed on the oil drop and slid through using a pair of tweezers.
- Immobilization of the protein (Figure 7)
- Pass Buffer 1 (Coupling Buffer) over the modified gold disk. This hydrates the gold surface and stabilizes the thiol layer (Recipe 10).
- In the 384-well microplate, dispense out all the solutions required for the immobilization procedure into the wells of the column 1 as marked in Figure 7.
- Add 80 μl EDC (400 mM) in wells 1A and 1B, followed by 80 μl of NHS (100 mM) in both the wells 1C and 1D.
- Leave 1E and 1F blank for mixing of EDC and NHS. This mixture will activate the gold sensor disk for ligand protein immobilization.
- Add 80 μl of the ligand protein (example: 5 mg/ml of EhP3) in both 1G and 1H wells.
- Finally, add 80 μl of Ethanolamine (1 M) in 1I and 1J, to be used for blocking the remaining unbound area on the sensor.
- Place the 384-well plate onto the rack as shown in Figure 1.
- Use the following steps for setting the data acquisition times in the Data Acquisition Software:
Steps Time Baseline (Coupling buffer) 120 s Activation (EDC/NHS) 300 s Coupling (Ligand protein) 900 s Blocking (Ethanolamine) 600 s Regeneration (coupling buffer) 120 s 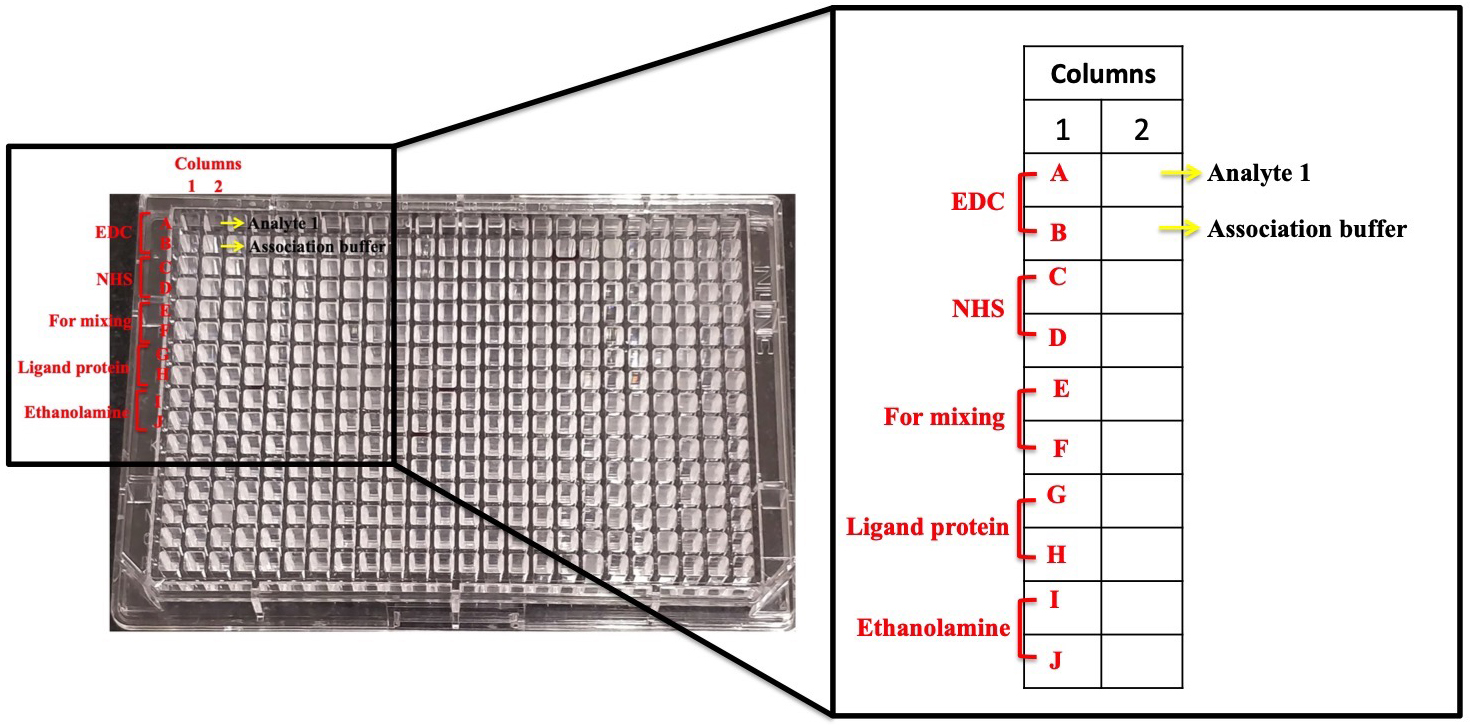
Figure 7. 384-well plate used in this protocol. Each well used in the plate is marked here.
- Interaction studies
- In column 2 of the 384-well microplate, add different dilutions of the Analyte, i.e., protein (or different proteins) to be used for determining their association kinetics.
- In well 2A, add 80 μl of Analyte 1 (example: Filamin) [or one dilution (1 μM) of a single protein (example: EhCoactosin)] followed by 80 μl of Buffer 2 (Association Buffer) (Recipe 11) in well 2B as reference.
- Similarly, add 80 μl of all the Analytes (examples: Actophorin, Profilin A, Profilin B, Twinfilin, EhCoactosin) or (examples: 2 μM, 4 μM, 8 μM, 10 μM of EhCoactosin) into alternative wells, followed by Buffer 2 (Association buffer) in each of the consecutive wells.
- Use Buffer 2 as the running buffer for the assay.
- Use the following incubation times for interaction studies:
Steps Time Baseline 120 s Association 300 s Dissociation 300 s Regeneration 120 s - The SPR assays also use controls like any other protein-protein interaction study. Bovine Serum Albumin generally serves as the negative control, i.e., unless the protein in focus binds to it specifically. BSA normally does not bind majority of the proteins. Similarly, if any known interacting protein is known, then it can be used as the positive control. Further, the buffers used in the study should be compatible to all the proteins. This can be checked by diluting a small amount of the proteins in all the buffers to be used, and check them for any precipitation.
Data analysis
Use the AutoLab Kinetic Evaluation software to obtain the ‘Differential curves’ (Figure 8). Use these to determine the Kon and Koff, along with with the affinity constant KD. The strength of interaction between two molecules is said to be stronger if the display a smaller KD value. The higher value of KD signifies weaker attraction between the interacting molecules. Kon represents the association constant with units in min-1 multiplied by concentration-1. Koff is the dissociation constant with units in min-1. Affinity constant is determined by the ratio of Koff/Kon in Molar units.
The Video 1 can be seen for data analysis steps.
- Open the Kinetic Evaluation Software.
- Follow the steps shown in the Video 1.Video 1. Steps to use the Kinetic Evaluation Software
- Select the folder where the files from the Data Acquisition software have been saved.
- In the folder, select all the series of injections recorded for the protein interaction study.
- On the left hand top corner, under the heading ‘Create Files’, click New Overlay and select all the injections to be used for the kinetic analysis.
- After this click on Signal Processing tab on the left hand bottom corner. Click on zoom and select the region of the sensogram before the association curve. Select Synchronize under the drop down tab of ‘Actions’. Next, click on ‘Normalize’ under the same tab and apply.
- To remove the regeneration curves, click on ‘Delete Selection’ and select the region after the dissociation curve.
- Again click on Normalize.
- Close the tab, and open New project under ‘Create Files’ and save the data with appropriate name.
- Next, open the Analysis wizard, select Equilibrium, click next. After this feed in the respective concentration.
- Finally, click next and the calculations for the kinetic parameters can be performed successfully.
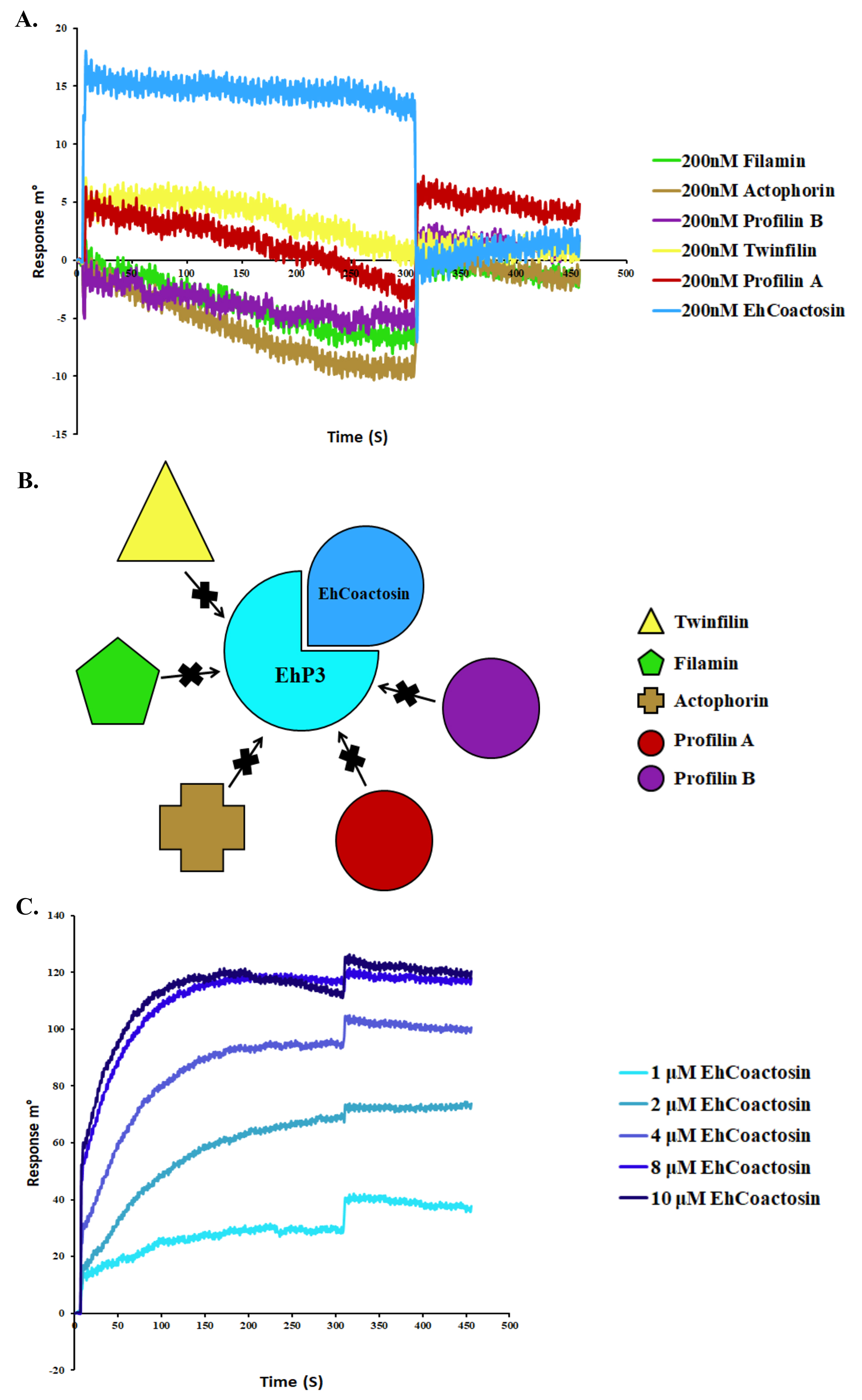
Figure 8. SPR analyses of cytoskeletal proteins to immobilized EhP3. A. Surface Plasmon Resonance was used to determine the interaction between EhP3 and cytoskeletal proteins like Filamin, Actophorin, Profilin A and B, Twinfilin, and EhCoactosin. EhP3 was immobilized covalently through amide coupling onto the gold sensor. Shown here are the sensorgrams of various proteins injected at 200 nM concentrations. B. Based on the sensograms observed in (A), we have depicted the interaction in this cartoon model, in which, only EhCoactosin showed binding to EhP3. The other proteins could not bind to the immobilized protein EhP3. C. Shown here are sensorgrams of EhCoactosin injections of increasing concentration. The equilibrium binding curves were used to derive the affinity constant KD. The concentrations of recombinant His6-Ehcoactosin are indicated alongside the binding curves
Notes
- The ideal buffer for proteins to be used for SPR studies would be HEPES, but if the protein stability is an issue, then keep the concentration of any other buffer to a maximum of 50 mM.
- EDC and NHS should be preferably freshly prepared as they are not very stable in solution. If prepared in larger quantity, one should store them as 300 μl aliquots in -20 °C for as long as thawed next time. However, it is not recommended to store them after thawing once.
- Preparation of the ligand in acetate buffer should always be done fresh. Moreover, the same acetate buffer stock should be maintained during the entire immobilization procedure.
- The pH of the coupling buffer can vary in case of different proteins. A general equation can be used to determine the pH of the coupling buffer: pH (buffer) = pI (ligand protein) - 0.5. pI is the isoelectric pH of the protein at which all its negative charges are equal to all its positive charges, making it electrically neutral. It can be theoretically calculated for any protein with known amino acid composition.
- There exists a linear relationship between the shift in SPR angle with the amount of the bound material. 120 m° change represents a change in surface protein mass of ~1 ng/mm2.
- Most kinds of organic solvents are not recommended for this machine, kindly refrain from using those in any of the sample preparations.
- Acetate buffer, with its lowest refractive index, serves as the best buffer for the immobilization process, as it gives the smallest SPR angle. Ethanolamine solution records the largest SPR angle.
- For any new protein-protein interaction, normally one can decide the concentration by referring to the available data on those proteins, if any. Otherwise, one can begin by taking a wide range of concentrations, example 50 nM-500 nM. If this range gives no information, one can increase the range up to 2 μM. Based on these reaction curves, ideal concentrations can be chosen for the particular set of proteins.
- Maintenance of the instrument:
- Hemi-cylinder should be regularly cleaned due to inevitable spillage of the immersion oil. As shown in Figure 2, M3X3 screw should be carefully removed and the hemi-cylinder should be cleaned with either ethanol or in an ultrasonic water bath. To dry the apparatus, ONLY lens tissue must be used. After the cleaning and drying, the hemi-cylinder must be placed and screwed back on the slider.
- In order to avoid any evaporation of solution from the cuvette, cover it with Para film.
- Always clean the instrument with distilled water. Use of any kind of buffer will lead to salt deposition and thus rendering the coating useless.
- The syringe pump seals should be changed on a yearly basis for optimum performance of the instrument.
Recipes
- 11-MUA solution
11 mg mercaptoundecanoic acid
50 ml ethanol
Filter sterilize the solution for optimum results - 400 mM EDC (2 ml)
153.4 mg EDC
2 ml demi water - 100 mM NHS
23 mg NHS
2 ml demi water - 1 M Ethanolamine
0.97 g salt
9 ml demi water
Use HCl to adjust its pH to 8.5
Finally, make up the volume to 10 ml - 1 M Acetate buffer pH 4.5
1.36 g salt
9 ml demi water
Use acetic acid to adjust the pH to 4.5
Finally, make up the volume to 10 ml (Abdul Rehman et al., 2013) - 1 M HEPES buffer pH 7.5
26.03 g salt
80 ml demi water
Use HCl to adjust the pH to 7.5
Finally, make up the volume to 100 ml - 5 M NaCl
29.22 g salt
100 ml demi water - 1 M NaOH
0.4 g of NaOH
10 ml demi water - 10% Tween 20
Prepared with demi water - Buffer 1 (Coupling buffer)
Dilute 0.5 ml of Acetate buffer stock to 50 ml solution using demi water - Buffer 2 (Association buffer) (1 L)
10 ml 1 M HEPES buffer pH 7.5
30 ml NaCl from its stock solution
0.5 ml 10% Tween 20
Make up the final volume to 1 L with demi water
Filter and use - Buffer 3 (Regeneration buffer)
Take 2.5 ml from NaOH stock solution
Make up the volum - Protein solutions
- Convert mg/ml protein concentration to molarity. Use the following formula:
μM of protein = (μg/μl of protein)/(MW in Da) - Next, dilute the protein stock in Buffer 2 to prepare 100 μM of working stock.
- Use the following calculations to prepare the dilution in Buffer 2 (Association buffer):
Sample name Volume of protein working stock Volume of Buffer 2 Dilution 1 (1 μM) 1 μl 99 μl Dilution 2 (2 μM) 2 μl 98 μl Dilution 3 (4 μM) 4 μl 96 μl Dilution 4 (8 μM) 8 μl 92 μl Dilution 5 (10 μM) 10 μl 90 μl - When using different proteins against one protein, use the same dilution (made in Buffer 2) for all the proteins. Make 1 μM dilution of all the proteins to be used as working stock. Take 20 μl of this stock and add 80 μl of Buffer 2 to prepare 200 nM solutions.
Protein name Concentration Analyte 1 (example: EhCoactosin) 200 nM Analyte 2 (example: Profilin A) 200 nM Analyte 3 (example: Profilin B) 200 nM Analyte 4 (example: Filamin) 200 nM Analyte 5 (example: Twinfilin) 200 nM Analyte 6 (example: Actophorin) 200 nM
- Convert mg/ml protein concentration to molarity. Use the following formula:
Acknowledgments
We acknowledge Advanced Instrument Research Facility, JNU and Manu Vashistha for SPR studies. We would also like to thank Prof. Alok Bhattacharya (Ashoka University, New Delhi, India) and Prof. Samudrala Gourinath (Jawaharlal Nehru University, New Delhi, India) for providing us the necessary help during the study. SA thanks the Department of Science and Technology for DST Inspire Faculty Award and Grant. PPR thanks the Council of Scientific and Industrial Research (CSIR) for SRF Fellowship.
Competing interests
The authors have declared that no competing interests exist.
References
- Abdul Rehman, S. A., Verma, V., Mazumder, M., Dhar, S. K. and Gourinath, S. (2013). Crystal structure and mode of helicase binding of the C-terminal domain of primase from Helicobacter pylori. J Bacteriol 195(12): 2826-2838.
- Agarwal, S., Anand, G., Sharma, S., Parimita Rath, P., Gourinath, S. and Bhattacharya, A. (2019). EhP3, a homolog of 14-3-3 family of protein participates in actin reorganization and phagocytosis in Entamoeba histolytica. PLoS Pathog 15(5): e1007789.
- Davis, T. M. and Wilson, W. D. (2000). Determination of the Refractive Index Increments of Small Molecules for Correction of Surface Plasmon Resonance Data. Anal Biochem 284: 348-353.
- Kumar, N., Somlata, Mazumder, M., Dutta, P., Maiti, S. and Gourinath, S. (2014). EhCoactosin stabilizes actin filaments in the protist parasite Entamoeba histolytica. PLoS Pathog 10(9): e1004362.
- Nguyen, H. H., Park, J., Kang, S. and Kim, M. (2015). Surface plasmon resonance: a versatile technique for biosensor applications. Sensors (Basel) 15(5): 10481-10510.
Article Information
Copyright
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Rath, P. P., Anand, G. and Agarwal, S. (2020). Surface Plasmon Resonance Analysis of the Protein-protein Binding Specificity Using Autolab ESPIRIT. Bio-protocol 10(4): e3519. DOI: 10.21769/BioProtoc.3519.
Category
Molecular Biology > Protein > Protein-protein interaction
Biophysics > Biophotonics
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link