- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Identification of Target Protein for Bio-active Small Molecule Using Photo-cross Linked Beads and MALDI-TOF Mass Spectrometry
Published: Vol 10, Iss 3, Feb 5, 2020 DOI: 10.21769/BioProtoc.3517 Views: 5838
Reviewed by: Gal HaimovichAlexandros C KokotosMartin V Kolev

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

ZnCl2 Precipitation-Assisted Sample Preparation for Proteomic Analysis
Qiqing He [...] Fuchu He
Jul 20, 2025 2763 Views

Protocol for the Preparation of a Recombinant Treacle Fragment for Liquid–Liquid Phase Separation (LLPS) Assays
Nadezhda V. Petrova [...] Artem K. Velichko
Sep 20, 2025 1858 Views

Optimized Secretome Sample Preparation From High Volume Cell Culture Media for LC–MS/MS Proteomic Analysis
Basil Baby Mattamana [...] Peter Allen Faull
Dec 20, 2025 1318 Views
Abstract
Development of methods for protein identification is one of the important aspects of proteomics. Here, we report a protocol for the preparation of compound conjugated beads by photo-crosslinking, affinity purification, gel electrophoresis, and highly sensitive mass spectrometric assay for drug-target identification. Although there are several other methods used for drug-target identification, such as biochemical fractionation or radioactive ligand binding assay, affinity purification is widely used for its straight-forward and easy approach. To identify the target protein of an inhibitor of cancer cell-accelerated fibroblast migration, we prepared the inhibitor-conjugated beads by photo-crosslinking. Proteins were pulled down from cell lysates by the compound beads and separated by SDS-PAGE, and a specifically pulled down protein was cut out, trypsin-digested, analyzed using matrix-assisted laser desorption ionization/time of flight mass spectrometry (MALDI-TOF-MS) and identified by peptide mass fingerprinting (PMF) method. Since the photo-crosslinking enables the immobilization of ligands on an affinity matrix in a functional group-independent manner, we do not have to determine the functional group of the compound to conjugate the matrix. In addition, as compared to other MS techniques such as electrospray ionization, MALDI offers a less complex sample preparation procedure and higher sensitivity, and thus is better suited for the rapid identification of proteins isolated by gel electrophoresis.
Keywords: ProteinBackground
The field of chemical proteomics is of great importance to drug discovery research enabling the identification and validation of a drug target and understanding the side effects as well as designing of more specific drugs. In our research, we used photo-crosslinked small molecule affinity matrix as a tool for identification of target protein of compound NPD8733, an inhibitor of cancer cell-accelerated fibroblast migration. The sepharose beads conjugated with NPD8733 and its control compound without activity were prepared by the photo-crosslinking technique (Kanoh et al., 2005; Suvarna et al., 2019). The proteins pulled down by the beads were separated using standard electrophoresis method. The protein pulled-down specifically by NPD8733-conjugated beads was cut out, subjected to in-gel trypsin digestion, and identified to be valosin-containing protein (VCP) using MALDI-TOF-MS and PMF analysis (Suvarna et al., 2019).
Pull-down assay using affinity beads is a commonly used method for identification of direct targets of small bioactive molecules. The preparation of affinity beads usually requires analysis of structure-activity relationship (SAR) so that the sites that do not affect the activity of the biomolecule can be identified and used for the attachment of an affinity tag such as biotin. Photo-affinity beads developed by Kanoh et al. overcome these drawbacks (Kanoh et al., 2005). The affinity beads prepared by photo-crosslinking technique, do not require SAR studies, since the photo-crosslinking technique utilizes a photo-generated carbene species to immobilize the small molecule on an affinity bead in a chemo- and site-nonselective manner. This unbiased nature of the photo-crosslinking technique proves beneficial for identifying the binding partners of unreactive small molecules; however it cannot be used for the small molecules that degrade upon UV irradiation (Kanoh et al., 2005).
The pulled-down proteins can be separated from the beads using electrophoretic protein separation under denaturing conditions or can also be directly digested on-beads using proteolytic enzymes. The electrophoretic method is inexpensive, easy to use, and has a wide separation range, making relative quantification easy. The separated proteins can be stained using Coomassie blue staining or silver staining methods. The resulting mixture of peptides can be analyzed using MS techniques such as MALDI-TOF or ESI-MS. When compared to ESI, MALDI-TOF is sensitive, requiring low picomole-level of gel-separated proteins. The sample preparation in MALDI is simple and thus suitable for rapid protein identification (Lubec and Afjehi-Sadat, 2007).
The photo-crosslinking method can also be applied for chemical arrays that can be used for ligand screening of a protein of interest. Since small molecules can be fixed in a functional group-independent manner, glass slides with thousands of compounds can be prepared without the need for SAR analysis, also making it possible to study SAR of compounds in a high-throughput manner as well (Kanoh et al., 2006).
Materials and Reagents
- 50 ml Falcon tubes
- Cloth
- Eppendorf tubes
- Micropipette loading tips for pull-down assay (Sorenson, catalog number: 13791)
- 15 cm Petri-dish for cell culture (Iwaki, catalog number: 3030-150)
- Glass vials (Maruemu Corp., No.2, catalog number: 5-098-04), 6 ml
- NHS beads (GE Healthcare, NHS-activated Sepharose 4 Fast Flow 25 ml, catalog number: 17090601)
- Photoaffinity PEG linker (Kondoh et al., 2015)
- Test compound (any vendor). In addition to the test compound, structurally similar and inactive compound was used as the negative control. We have frequently experienced that negative compound conjugated beads act as better “negative control beads” when compared to the naked beads
- Spin column (GeneDesign, Inc., EconoSpin Empty Column, catalog number: EP-31301)
- Mouse embryonic fibroblast cell line NIH3T3 (obtained from RIKEN Cell Bank, catalog number: RCB1862)
- Human breast cancer cell line MCF7 (obtained from RIKEN Cell Bank, catalog number: RCB1904)
- Dulbecco's Modified Eagle Medium (D-MEM) media (Gibco, catalog number: C11995500BT)
- Fetal calf serum (FCS) (Nichirei Bioscience Inc. Tokyo, Japan) used at 10% in DMEM
- Penicillin and Streptomycin (Pen Strep, Gibco, catalog number: 15140-122) used at 100x dilution in DMEM
- HEPES (Sigma, catalog number: H3375)
- HCl (FUJIFILM Wako Pure Chemical, catalog number: 080-01066)
- NaCl (FUJIFILM Wako Pure Chemical, catalog number: 191-01665)
- KCl (FUJIFILM Wako Pure Chemical, catalog number: 163-03545)
- Na2HPO4·12H2O (FUJIFILM Wako Pure Chemical, catalog number: 196-02835)
- KH2PO4 (FUJIFILM Wako Pure Chemical, catalog number: 169-04245)
- Dioxane (FUJIFILM Wako Pure Chemical, catalog number: 042-03766)
- MQ-water (Purified water produced by MilliQ Q-POD, Millipore)
- NaN3 (FUJIFILM Wako Pure Chemical, catalog number: 199-11095)
- EDTA (FUJIFILM Wako Pure Chemical, catalog number: 345-01865)
- Sequencing Grade Modified Trypsin (Promega, catalog number: V5111)
- Acetonitrile (Nacalai tesque, catalog number: 00420-41)
- TFA (FUJIFILM Wako Pure Chemical, catalog number: 201-06393)
- Methanol (FUJIFILM Wako Pure Chemical, catalog number: 131-01826)
- α-Cyano-4-hydroxycinnamic acid (FUJIFILM Wako Pure Chemical, catalog number: 037-19261)
- NaHCO3 (FUJIFILM Wako Pure Chemical, catalog number: 191-01305)
- NH4HCO3 (FUJIFILM Wako Pure Chemical, catalog number: 017-02875)
- Ethanolamine (FUJIFILM Wako Pure Chemical, catalog number: 012-12455)
- Tris (FUJIFILM Wako Pure Chemical, catalog number: 201-06273)
- DTT (FUJIFILM Wako Pure Chemical, catalog number: 042-29222)
- Iodoacetamide (FUJIFILM Wako Pure Chemical, catalog number: 093-02892)
- Coomassie Brilliant blue R250 (CBB) (FUJIFILM Wako Pure Chemical, catalog number: 031-17922)
- Acetic acid (FUJIFILM Wako Pure Chemical, catalog number: 017-00256)
- Isopropanol (FUJIFILM Wako Pure Chemical, catalog number: 325-00045)
- Sodium Dodecyl Sulfate (SDS) (FUJIFILM Wako Pure Chemical, catalog number: 192-08672)
- Bromophenol blue (FUJIFILM Wako Pure Chemical, catalog number: 021-02911)
- Glycerol (FUJIFILM Wako Pure Chemical, catalog number: 075-00611)
- 2-Mercaptoethanol (FUJIFILM Wako Pure Chemical, catalog number: 28625.01)
- Ammonium Persulfate (APS) (FUJIFILM Wako Pure Chemical, catalog number: 802811)
- N,N,N’,N’-tetramethylethylenediamine (TEMED) (FUJIFILM Wako Pure Chemical, catalog number: 202-04003)
- Razor (FEATHER, catalog number: 99077)
- cOmplete EDTA-free Protease inhibitor cocktail tablet (Roche diagnostic, Mannheim, Germany, catalog number: 11836170001)
- Pierce BCA protein assay kit (Thermo Fisher Scientific, catalog number: 23225)
- Wash/binding buffer containing protease inhibitors (see Recipes)
- 1x SDS buffer (see Recipes)
- Binding/wash buffer (see Recipes)
- aq. HCl (see Recipes)
- PBS (see Recipes)
- Coupling buffer (see Recipes)
- Blocking solution (see Recipes)
- Decolorant (see Recipes)
- Reduction solution (see Recipes)
- Alkylating solution (see Recipes)
- Digestion solution (see Recipes)
- Extraction solution 1 (see Recipes)
- Extraction solution 2 (see Recipes)
- Matrix solution (see Recipes)
- Staining solution (see Recipes)
- Destaining solution (see Recipes)
- 10x Tris-glycine running buffer (see Recipes)
- 5% Stacking gel (see Recipes)
- 10% Resolving gel recipe (see Recipes)
Equipment
- UV crosslinker (UVP, Ultraviolet Crosslinker, CL-1000)
- LCMS (LC: Waters, ACUITY UPLC H-class, MS: SCIEX, API 3200 LC-MS/MS)
- Agilent Oven (Agilent Technologies, Hybridization Oven, G2545A)
- High-performance rotary evaporator (Scrum, Genevac, EZ-2 plus)
- Rotator (TAITEC, model: RT-50)
- Desk-top centrifuge machine (Corning, model: 6765)
- Standard refrigerated centrifuge for 15/50 ml and Eppendorf tubes (Tomy, model: MX-301)
- Savant SpeedVac Concentrator (ThermoFisher Scientific, model: SVC100H)
- Heat block (EYELA, model: MG-1200)
- Ultrasonic bath (AS ONE, US CLEANER US-1R)
- Sonicator (Tomy, model: UR-20P)
- Slab type SDS-electrophoresis equipment (NIHON-EIDO, model: NA-1010)
- MALDI-TOF mass spectrometer (Ultraflex, Bruker Daltonik GmbH)
Software
- Biotools (Biotools version 3.2, Bruker Daltonik GmbH, https://www.bruker.com/)
- Mascot search program (Mascot Server, Matrix Science Inc, http://www.matrixscience.com)
Procedure
- UV stability check of compound
- Put small amount (ca. 0.1-0.3 mg) of compound in glass vial.
- Dissolve compound with 20 μl of methanol (or soluble solvent with relatively low boiling point) and divide it into two vials
- Concentrate and dry in vacuo with a high-performance rotary evaporator (Genevac) at 35 °C by “Med + Low BP mixture” solvent mode for 1 h.
- Irradiate one of the two vials with UV (365 nm; 4 J/cm2) with UV crosslinker.
- Dissolve both UV(+) and UV(-) compounds with methanol and analyze with LCMS.
- Compare LCMS chart of UV(+) and UV(-) compounds and check the stability of the compound against UV irradiation.
- Compound beads preparation
- Wash 400 μl of NHS beads with 400 μl of 1 mM aq. HCl 3 times and subsequently with 400 μl of coupling buffer 3 times at room temperature. Briefly, add the washing solution, mix by pipetting using a pipet tip to suspend the beads totally, centrifuge in a desk-top centrifuge machine at 2,000 x g for one minute, remove and discard the supernatant.
- Dissolve 8.9 mg (18 μmol) of photoaffinity PEG linker in 400 μl of coupling buffer and add to the beads.
- Incubate the beads suspension under rotation (10 rpm) in Agilent Oven at 37 °C for 2 h.
- Wash beads with 400 μl of coupling buffer 3 times as with Step B1.
- Add 1 ml of blocking solution.
- Incubate beads suspension under rotation in Agilent Oven at 37 °C for 1 h.
- Wash beads with 400 μl of methanol 3 times and subsequently with 400 μl of MQ-water 3 times in a spin column. Briefly, add washing solution to the beads on the spin column, place the column in an Eppendorf tube, and centrifuge in a desk-top centrifuge machine at 2,000 x g for 1 min, following which the filtrate collected into the Eppendorf tube.
- Transfer the beads to a glass vial and add a methanol-based solution containing the compound (6 μmol/400 μl of methanol).
- Concentrate and dry in vacuo with high-performance rotary evaporator (Genevac) for 1 to 1.5 h (depends on solvent).
- Irradiate the beads with UV using UV crosslinker 4 J/cm2.
- Wash the free compounds with 400 μl of methanol (or soluble solvent) 15 times and subsequently with 400 μl of MQ-water 3 times in a spin column as in Step B7.
- Suspend the compound beads in 400 μl of PBS-1 mM NaN3 and store at 4 °C until use.
- Pull-down assay
- Preparation of cell lysates
- Co-culture NIH3T3 cells and MCF7 cells (5:1; 2 x 106 and 0.4 x 106/15 cm dish, respectively) in D-MEM with 10% FCS containing penicillin and streptomycin for 48 h and harvest, wash with PBS, resuspend in wash/binding buffer containing protease inhibitors. (We usually use approximately 2 x 108 cells from 20 of 15 cm dishes and resuspend the cells in 10 ml of wash/binding buffer in 50 ml Falcon tubes. Sonicate the sample for 3 min (30 s on and 30 s off). Place the Falcon tubes on ice during sonication.)
- Discard the insoluble material by centrifugation at 4,400 x g for 20 min at 4 °C and transfer the supernatant into fresh falcon tubes. Typically, the concentration of the lysate used was about 16 mg/ml and we used lysates containing about 8 mg of protein for pull-down with 30 µl compound beads. The protein estimation was done using Pierce BCA protein assay kit according to their instruction.
- Aliquot 30 µl beads (conjugated with test compounds, negative compounds and naked) using wide bore-tip into fresh Eppendorf tubes.
- Wash the beads by adding 300 µl wash/binding buffer, mix gently and spin down at maximum speed for 30 s. Discard the supernatant carefully. Repeat this step twice.
- Add lysate containing 8 mg of protein in 500 µl of wash/binding buffer to each of the Eppendorf tube containing the beads. (Perform all the steps at 4 °C)
- Incubate the mixture containing beads and cell lysate at 4 °C under gentle rotating condition using a rotating mixer for at least 3 h.
- Spin down and discard the supernatant carefully. Wash the beads by adding 500 µl wash/binding buffer, mix gently and spin down. Discard the supernatant carefully. Repeat this step at least 3 times.
- After the last wash, carefully discard all the supernatant. Try not to leave any supernatant and also be careful not to displace the beads. Use loading tips for removing the supernatant. You may move the beads into fresh Eppendorf tubes if required.
- Add 30 µl 1x SDS buffer to the beads, keep the Eppendorf tubes at 100 °C for 5 min.
- Allow the sample to cool down and then load the supernatant onto a 10% SDS-PAGE gel using SDS-electrophoresis equipment (NIHON-EIDO NA-1010) and keep it for overnight staining using CBB stain. The gel was then washed in destaining solution for at least two hours. An example of stained gel is shown in Figure 1. The analyzed region is shown.
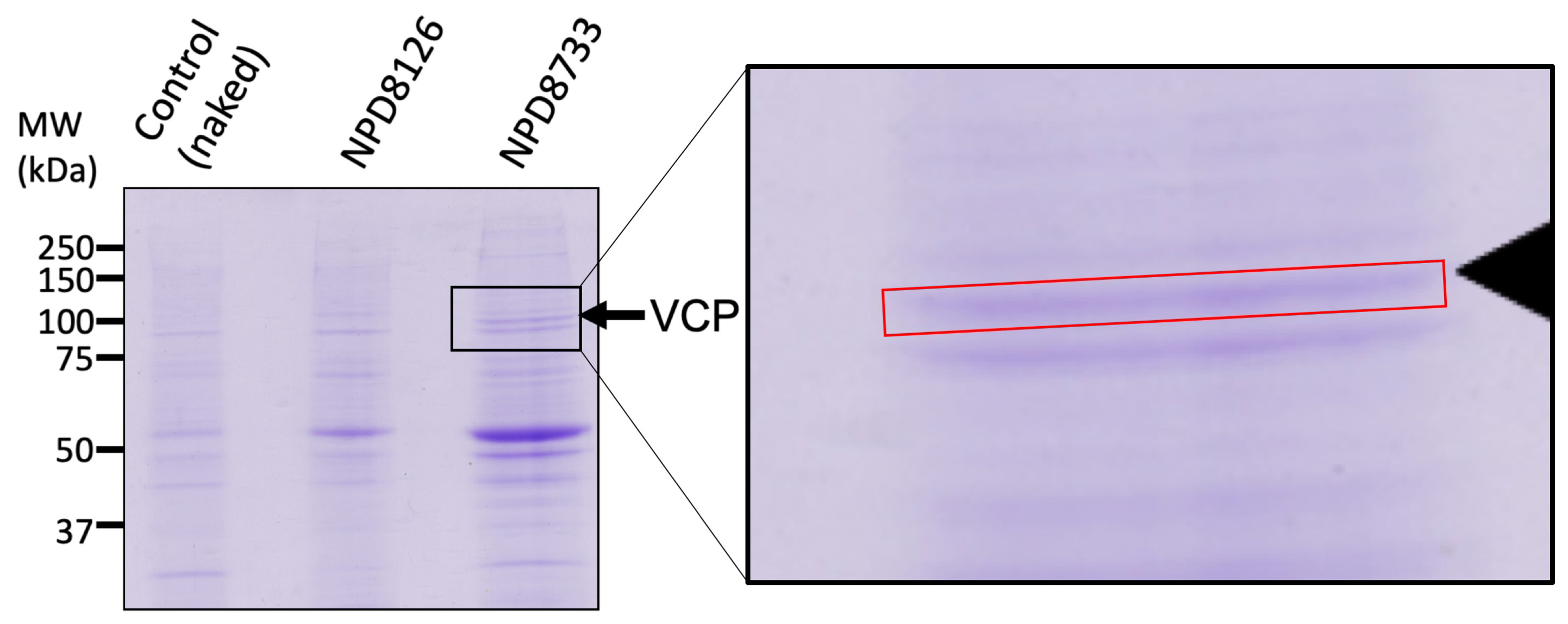
Figure 1. NIH3T3 and MCF7 cells were co-cultured at a 5:1 ratio and their cell lysates were incubated with control beads, NPD8126 beads, or NPD8733 beads for 3 h. The reacted beads were washed, and the eluted proteins were separated using SDS-PAGE and stained by CBB. The protein specifically co-precipitated with NPD8733 beads was identified by MALDI-TOF-MS to be VCP (arrow). The red rectangular box indicates the approximate region of the gel portion cut out for protein identification.
- Preparation of cell lysates
- Identification of binding proteins by peptide mass fingerprinting using MALDI-TOF mass spectrometry
- Cut the region of interest [usually 5 mm x 1 mm x 1 mm (thickness)] using a razor and transfer half of it (2.5 mm x 1 mm x 1 mm) into a clean Eppendorf tube. The other half is stored for backup.
- Wash the gel by adding 400 µl of water, incubate for 10 min and discard the supernatant. Repeat this step 5 times.
- Incubate with 100 µl of decolorant for 10 min at 37 °C.
- Wash the gel by adding 200 µl of acetonitrile, incubate for 10 min and discard the supernatant. Repeat this step twice.
- Dry in vacuo using SpeedVac concentrator at a machine-fixed rotation rate for about 45 min at room temperature (without heating). As dehydration proceeds, the gel usually becomes opaque.
- Add 50 µl of reduction solution to the dried gel and incubate at 56 °C for 30 min on a heat block.
- Add 100 µl of alkylating solution to the gel and incubate at 37 °C for 30 min covered with a cloth to protect from light.
- Wash the gel with 200 µl of 100 mM NH4HCO3 solution and discard the supernatant.
- Wash with 200 µl of acetonitrile twice for 10 min.
- Dry in vacuo using SpeedVac concentrator.
- Add 3 µl of digestion solution to the dried gel on ice for 30 min.
- Add 10 µl of 10 mM Tris-HCl (pH 8.8), and incubate at 37 °C overnight.
- Remove the supernatant to a new microtube. Add 10 µl of extraction solution 1 to the remaining gel bits and incubate for 10 min in an ultrasonic bath with ice-cold water at low power setting.
- Remove the supernatant and add it to the supernatant containing microtube from Step D13.
- Add 10 µl of extraction solution 2 to the remaining gel bits and incubate for 10 min in an ultrasonic bath with ice-cold water.
- Remove the supernatant into the supernatant containing microtube from Step D13.
- Repeat the Steps D15 and D16.
- Add 10 µl of acetonitrile to the remaining gel bits and incubate for 10 min in an ultrasonic bath with ice-cold water at low power setting.
- Remove the supernatant into the supernatant containing microtube from Step D13.
- Dry in vacuo using SpeedVac concentrator.
- Dissolve with 3 µl of 0.1% TFA including 5% acetonitrile.
- Mix 0.25 µl of the sample and equal volume of matrix solution on the target plate of a MALDI-TOF mass spectrometer.
- Analyze by operating a MALDI-TOF mass spectrometer in the positive ion reflector mode.
Data analysis
Perform protein identification by peptide mass finger printing using the Mascot search program (standard setting for trypsinized sample) based on the peak list generated by the software (Biotools) provided by the manufacturer (Bruker Daltonik GmbH).
Recipes
- 1x SDS buffer
0.0005% Bromophenol blue
10% Glycerol
2% SDS
63 mM Tris HCl pH 6.8
0.1% 2-Mercaptoethanol (Add before use) - Binding/wash buffer containing protease inhibitors
50 mM HEPES
150 mM NaCl
1 mM EDTA
Adjust pH 7.5 and volume up with MQ water
Store at 4 °C
Add protease inhibitor (complete EDTA free-protease cocktail tablets) immediately before use - aq. HCl
1 mM HCl - PBS
137 mM NaCl
2.7 mM KCl
10 mM Na2HPO4·12H2O
1.8 mM KH2PO4 - Coupling buffer
0.1 M aqueous NaHCO3:dioxane = 1:1 - Blocking solution
1 M ethanolamine in 0.1 M Tris-HCl pH 8.0 buffer - Decolorant
50% acetonitrile and 50 mM NH4HCO3 - Reduction solution
100 mM DTT and 100 mM NH4HCO3
Prepare fresh immediately prior to use - Alkylating solution
100 mM iodoacetamide and 100 mM NH4HCO3
Prepare fresh immediately prior to use - Digestion solution
25 µg/ml trypsin in 10 mM Tris-HCl pH 8.8 buffer - Extraction solution 1
0.05% TFA and 50% acetonitrile - Extraction solution 2
0.03% TFA and 66% acetonitrile - Matrix solution
α-Cyano-4-hydroxycinnamic acid (10 mg/ml) dissolved in 50% acetonitrile/0.1% TFA - Staining solution
0.3 g Coomassie Brilliant blue R250
250 ml isopropanol
300 ml acetic acid
650 ml MQ water - Destaining solution
300 ml methanol (30%)
100 ml acetic acid (10%)
600 ml Milli Q water - 10x Tris-glycine running buffer
25 mM Tris
192 mM glycine
0.1% SDS - 5% Stacking gel
Milli Q 1.4 ml 30% acrylamide mix 0.33 ml 1.0 M Tris (pH 6.8) 0.25 ml 10% SDS 0.02 ml 10% APS 0.02 ml TEMED 0.002 ml - 10% Resolving gel recipe
Milli Q 1.9 ml 30% acrylamide mix 1.7 ml 1.5 M Tris (pH 8.8) 1.3 ml 10% SDS 0.05 ml 10% APS 0.05 ml TEMED 0.002 ml
Acknowledgments
We thank the members of RIKEN NPDepo for providing the chemical libraries; Miho Tanaka (RIKEN), Dr. Takehiro Suzuki (RIKEN), and Dr. Naoshi Dohmae (RIKEN) for protein identification and Harumi Aono (RIKEN) and Emiko Sanada (RIKEN) for continuous technical help during the study. This research was partially supported by the research grant from JSPS KAKENHI (Grant Number JP16H06276, JP17H06412, JP17K07783, JP18H05503, JP18K05366, JP18H02555, JP19H05302) and AMED (Grant Number JP19cm0106112). The protocol for compound beads preparation was adapted from a previous work (Kondoh et al., 2015).
Competing interests
The authors declare no conflicts of interest.
References
- Kanoh, N., Honda, K., Simizu, S., Muroi, M. and Osada, H. (2005). Photo-cross-linked small-molecule affinity matrix for facilitating forward and reverse chemical genetics. Angew Chem Int Ed Engl 44(23): 3559-3562.
- Kanoh, N., Asami, A., Kawatani, M., Honda, K., Kumashiro, S., Takayama, H., Simizu, S., Amemiya, T., Kondoh, Y., Hatakeyama, S., Tsuganezawa, K., Utata, R., Tanaka, A., Yokoyama, S., Tashiro, H. and Osada, H. (2006). Photo-cross-linked small-molecule microarrays as chemical genomic tools for dissecting protein-ligand interactions. Chem Asian J 1(6): 789-797.
- Kondoh, Y., Honda, K. and Osada, H. (2015). Construction and application of a photo-cross-linked chemical array. Methods Mol Biol 1263: 29-41.
- Lubec, G. and Afjehi-Sadat, L. (2007). Limitations and pitfalls in protein identification by mass spectrometry. Chem Rev 107(8): 3568-3584.
- Suvarna, K., Honda, K., Muroi, M., Kondoh, Y., Osada, H. and Watanabe, N. (2019). A small-molecule ligand of valosin-containing protein/p97 inhibits cancer cell-accelerated fibroblast migration. J Biol Chem 294(9): 2988-2996.
Article Information
Copyright
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Suvarna, K., Honda, K., Muroi, M., Kondoh, Y., Watanabe, N. and Osada, H. (2020). Identification of Target Protein for Bio-active Small Molecule Using Photo-cross Linked Beads and MALDI-TOF Mass Spectrometry. Bio-protocol 10(3): e3517. DOI: 10.21769/BioProtoc.3517.
- Suvarna, K., Honda, K., Muroi, M., Kondoh, Y., Osada, H. and Watanabe, N. (2019). A small-molecule ligand of valosin-containing protein/p97 inhibits cancer cell-accelerated fibroblast migration. J Biol Chem 294(9): 2988-2996.
Category
Cancer Biology > Cancer biochemistry > Protein
Biochemistry > Protein > Isolation and purification
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










