- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Measurement of the Promoter Activity in Escherichia coli by Using a Luciferase Reporter
Published: Vol 10, Iss 2, Jan 20, 2020 DOI: 10.21769/BioProtoc.3500 Views: 5305
Reviewed by: Juan Facundo Rodriguez AyalaJose Antonio Reyes-DariasJan-Ulrik Dahl

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Development and Quantitation of Pseudomonas aeruginosa Biofilms after in vitro Cultivation in Flow-reactors
Yingdan Zhang [...] Haihua Liang
Aug 20, 2021 3680 Views

Quantification of RuBisCO Expression and Photosynthetic Oxygen Evolution in Cyanobacteria
Mateusz Kędzior and Betul Kacar
Oct 20, 2021 3334 Views

A Practical CRISPR-Based Method for Rapid Genome Editing in Caulobacter crescentus
Xuezhou Yuan [...] Jingxian Sun
Nov 5, 2025 1812 Views
Abstract
The reporter system is widely used technique for measuring promoter activity in bacterial cells. Until now, a number of reporter system have been developed, but the bioluminescent reporter constructed from the bacterial luciferase genes is one of the useful systems for measuring in vivo dynamics of gene expression. The introduced bioluciferase lux reporter enables easy, fast, and sensitive measurement of the promoter activity without cell lysis because the substrates of bioluminescent reaction are synthesized inside the bacterial cell, thereby allowing low-cost experiments. This protocol describes a high throughput technique to measure the promoter activity in Escherichia coli K-12 using the lux reporter system.
Keywords: lux operonBackground
The promoter activity in vivo was measured using a reporter system such as the lacZ (encoding β-galactosidase), gus (encoding β-gulucuronidase), and cat (encoding chloramphenicol acetyltransferase) genes. In the case of the lacZ reporter system, for instance, the test promoter sequence is fused to a promoter-less lacZ gene, creating a test promoter-lacZ fusion gene, which is then transferred into a recipient cell. For the measurement of the activity of the test promoter, however, the whole cell lysate must be prepared to detect in vitro β-galactosidase activity by adding a substrate such as ONPG (O-Nitrophenyl-β-D-galactopyranoside). To avoid such biochemical procedures, the fluorescent gfp gene, coding green fluorescent protein (GFP), was employed as a reporter which can be detected without cell lysis. Thus, the fluorescent reporter system is more convenient than the systems which requires measurement of enzymatic activity. However, the fluorescent proteins have a technical limitation especially in genes that are expressed at low levels because of high background noise that arises from intrinsic autofluorescence of cells. To overcome this problem, the luminescent reporter has been developed, which catalyzes bioluminescence reactions using the substrate as luciferins (Meighen, 1991). The Photorhabdus luminescens bioluminescence luxCDABE genes, coding two luciferase subunits (LuxAB) and three proteins (LuxCDE), which are important for substrate biosynthesis (Bjarnason et al., 2003). Once the test promoter is fused to promoter-less luxCDABE, both luciferase and its substrate are expressed under the control of the test promoter, and the promoter activity can be easily determined by measuring luminescence without the cell lysis (Bjarnason et al., 2003). This bioluminescent reporter system is recognized as a powerful high-throughput assay for studying continuous kinetics of promoter activity (Yamanaka et al., 2018; Burton et al., 2010). In this protocol, we describe how to construct the bioluminescent reporter system and how to measure the promoter activity in E. coli.
Materials and Reagents
- Pipette tips (Thermo Fisher Scientific, catalog number: QSP Liquid Handling Products 110-Q and 111-Q) (Gilson, catalog number: DIAMOND Tips DL10 and D5000)
- 1.5 ml plastic tube (Rikaken, catalog number: STAR MicroTestTube 1.5 ml RSV-MTT1.5)
- 0.22 μm filter (Advantec, catalog number: 13CP020AS)
- Glass tubes (Iwaki, catalog number: TEST18NP)
- Sterile 50 ml plastic tube (Iwaki, catalog number: 2345-050)
- BD Falcon 96-well plates, Black/Clear BD Optilux (Becton Dickinson, catalog number: 353948)
- 96-well white plate (Becton Dickinson, catalog number: 353377)
- Petri dishes (Rikaken, catalog number: STAR SDish9015 ver.2 RSU-SD9015-2)
- pLUX vector (Burton et al., 2010)
- Specific primers (Thermo Fisher Scientific, Custom DNA Oligos) (Table 1)
- TaKaRa Ex Taq (Takara Bio, catalog number: RR001A)
- NucleoSpin Gel and PCR Clean-up (Macherey-Nagel, catalog number: 740609.10)
- Restriction enzyme Xho I (Takara Bio, catalog number: 1094)
- Restriction enzyme Bam HI (Takara Bio, catalog number: 1010)
- In-Fusion HD Cloning Plus (Takara Bio, catalog number: 638920)
- Kanamycin Monosulfate (Nacalai Tesque, catalog number: 19839-44)
- Plasmid DNA Extraction Mini Kit (Favorgen, catalog number: FAPDE 001)
- BigDye® Terminator v3.1 Ready Reaction Mix (Applied Biosystems, catalog number:4337455)
- IPTG (Nacalai Tesque, catalog number: 06289-67)
- 3 M sodium acetate (Nacalai Tesque, catalog number: 06893-24)
- Ethanol (Nacalai Tesque, catalog number: 14710-25)
- Hi-DiTM Formamide (Applied Biosystems, catalog number: 4311320)
- BactoTM tryptone (BD Biosciences, catalog number: 211705)
- BactoTM yeast extract (BD Biosciences, catalog number: 212750),
- NaCl (Nacalai Tesque, catalog number: 31320-05)
- NaOH (Nacalai Tesque, catalog number: 31511-05)
- Na2HPO4·12H2O (Nacalai Tesque, catalog number: 31722-45)
- KH2PO4 (Wako, catalog number: 498748161612)
- MgCl2·6H2O (Nacalai Tesque, catalog number: 20908-65)
- K2SO4·12H2O (Nacalai Tesque, catalog number: 01727-25)
- NH4Cl2 (Nacalai Tesque, catalog number: 02424-55)
- CaCl2 (Nacalai Tesque, catalog number: 08894-25)
- D-(+)-Glucose (Nacalai Tesque, catalog number: 16805-35)
- 0.5 M EDTA (Nacalai Tesque, catalog number: 06894-14)
- Competent E. coli DH5α, provided from National Institute of Genetics in Japan (preparation at time of use) (see Recipes)
- 50 mg/ml kanamycin (see Recipes)
- LB broth (see Recipes)
- LB agar with 50 μg/ml kanamycin (see Recipes)
- 125 mM EDTA (see Recipes)
- 70% ethanol (see Recipes)
- M9-Glucose medium (see Recipes)
Equipment
- Pipettes (Gilson, models: PIPETMAN P2, P10, P20, P100, P200, P1000, P5000)
- Centrifuge (Tomy, model: MX-301)
- Thermal Cycler (Applied Biosystems, model: 2720Thermal Cycler)
- Temperature chamber (Taitec, model: Thermo minder SM-10R)
- Water bath shaker (Taitec, model: Personal-11)
- DNA sequencer (Applied Biosystems, model: 3500Genetyc Analyzer)
- Plate reader (Corona, model: MTP−880Lab)
- Autoclave (Tomy Seiko, model: LSX-500)
Software
- SF6 for Windows (Corona, in only Japanese)
- Microsoft Excel (Microsoft)
Procedure
- Construction of pLUX reporter plasmids
- Determine promoter region around a target gene. Typically, we take a region of DNA from 500 base-pairs (bp) upstream to 150 bp downstream of distal transcription start site (see for instance, the determined promoters of 18 target genes Figure 1).
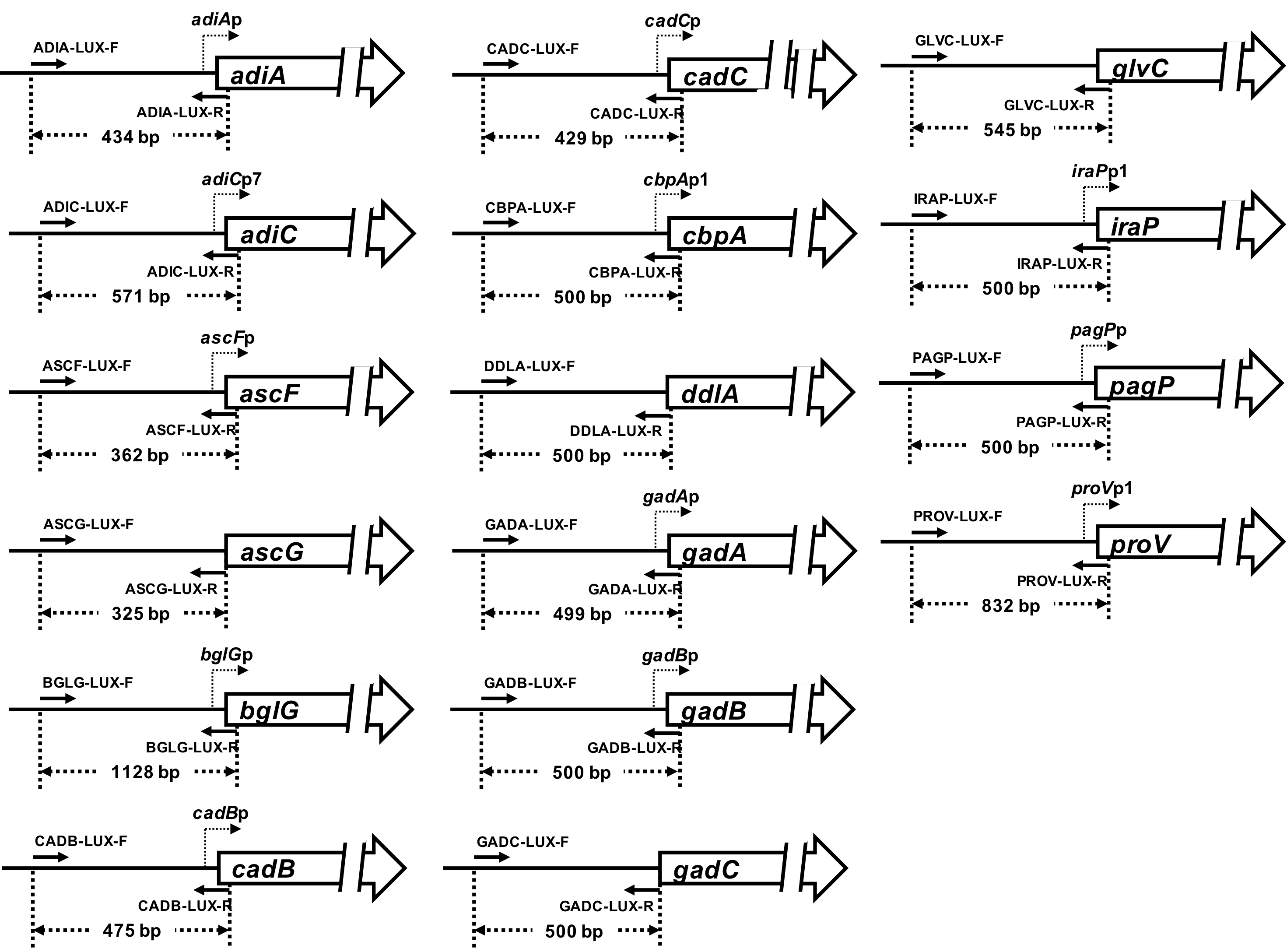
Figure 1. Eighteen E. coli promoter regions into pLux - Design primers which including 15 overlapping nucleotides (nt) with the pLUX vector for amplification of the determined promoter region (see Figure 2 and Table 1).
- Amplify target promoter regions by PCR using Ex Taq polymerase (Takara Bio). Standard reaction condition is used as described in the manual for Ex Taq polymerase (Takara Bio). In most of the cases, thermal cycle conditions are as follows: after heating at 98 °C for 5 min, process 30 cycles of 98 °C for 10 s (denature), 50 °C for 30 s (annealing), and 72 °C for 1 min (elongation) (Figure 2).
- Purify the PCR product by NucleoSpin Gel and PCR Clean-up (Macherey-Nagel).
Note: When PCR product includes non-specific bands, the target product is cut from agarose gel after electrophoresis with total volume of PCR product and then purified by NucleoSpin Gel and PCR Clean-up (Macherey-Nagel). - Digest pLUX vector using restriction enzymes Xho I and Bam HI, and then purify the liner pLUX by NucleoSpin Gel and PCR Clean-up (Macherey-Nagel).
Note: For cloning into pLUX, we recommend the double-digestion of pLUX by both Xho I and Bam HI (Figure 2). - Clone a target promoter as the purified PCR product into the liner pLUX using In-Fusion HD Cloning Plus (Takara Bio).
Note: We recommend in vivo E. coli cloning (iVEC) as an alternative DNA cloning method (Nozaki et al., 2019), which is provided from National BioResource Project (NBRP) of Japan. - Add a part (~10 μl) of the In-Fusion reaction mixture into 0.1 ml of the suspension of competent E. coli DH5α (see Recipes), and then incubate the mixture on ice for 30 min.
- Heat the mixture at 42 °C for 45 s, and immediately add 0.9 ml of LB broth to the mixture.
- Incubate 1 ml of the suspension of E. coli DH5α transformants at 37 °C for 60 min.
- Spread 0.1-0.2 ml of the suspension of E. coli DH5α transformants onto the LB agar with 50 μg/ml kanamycin, and then incubate the agar plate at 37 °C overnight.
- Isolate a transformant of E. coli DH5α harboring the cloned candidates as a single colony on the LB agar with 50 μg/ml kanamycin.
- Inoculate a single colony into 5 ml of LB broth containing 0.05 ml of 50 mg/ml kanamycin in a glass tube (Iwaki), and incubate the culture in water bath (Taitec) at 37 °C with shaking (120 rpm) overnight.
- Isolate the plasmid from the transformant cells of 5 ml overnight culture by Plasmid DNA Extraction Mini Kit (Favorgen).
- Sanger reaction is performed with the isolated plasmid as a template, LUX-R primer (Table 1), and BigDye® Terminator v3.1 Ready Reaction Mix (Applied Biosystems) according to the recommended procedure by Supplier.
- Transfer 20 μl of sanger product into a sterile 1.5 ml plastic tube and then add 2 μl of 125 mM EDTA, 2 μl of 3 M sodium acetate, and 50 μl of ethanol (Nacalai Tesque).
- Mix the solution by vortex mixer for 15 s.
- Collect DNA pellet by centrifugation (17,800 x g, 4 °C, 15 min).
- Add 70 μl of 70% ethanol and collect DNA pellet by centrifugation (17,800 x g, 4 °C, 15 min).
- Dissolve DNA pellet by 15 μl of Hi-DiTM Formamide (Applied Biosystems).
- Determine DNA sequence of the promoter region cloned into the isolated plasmid with DNA sequencer (Applied Biosystems).

Figure 2. Strategy for construction of pLUX reporter plasmid. The promoter region was amplified by PCR using the E. coli K-12 genome as the template and a pair of specific primers (Table 1). The amplified DNA fragment was inserted into a pLUX vector using the In-Fusion HD cloning kit (Takara Bio). After transformation of DH5α, the plasmid was purified from a culture of transformant cells. The DNA sequence of insertion on the resulting plasmids was confirmed by DNA sequencing.
Table 1. The used primers and pLUX derivatives
The plasmids constructed in this study could be provided from National BioResource Project (NBRP) E. coli of Japan.
- Determine promoter region around a target gene. Typically, we take a region of DNA from 500 base-pairs (bp) upstream to 150 bp downstream of distal transcription start site (see for instance, the determined promoters of 18 target genes Figure 1).
- Measurement of luciferase activity in E. coli
- Transform E. coli strains by the cloned luciferase reporter plasmids.
Note: The ∆hns∆hha∆ydgT strain, isolated from E. coli K-12 W3110 strain, deleted three genes, hns, hha, and ydgT, in its genome (Ueda et al., 2013). H-NS plays a role in transcriptional silencing of genes, which is modulated by Hha and YdgT proteins in E. coli. The ∆hns∆hha∆ydgT strain was transformed with pQE80L, pQE80Lhns which carries hns gene in pQE80L, pQE80Lhns-I70A which carries hns gene with substitutions of Ile70Ala, or pQE80Lhns-L75A which carries hns gene with substitutions of Leu75Ala. These transformants were used as hosts for luciferase measurements. - Inoculate at least three single colonies of E. coli transformant in glass tubes separately under M9-glucose medium including 50 μg/ml kanamycin.
Note: In addition of kanamycin, ampicillin should be added in medium at the final concentration of 100 μg/ml for the ∆hns∆hha∆ydgT strain harboring a pQE80L derivative and a pLux derivative. - Incubate the pre-cultures at 37 °C with shaking for overnight.
- Inoculate 100 μl of overnight pre-culture in 10 ml of fresh M9-glucose medium including 50 μg/ ml kanamycin.
- Incubate the 10 ml cultures at 37 °C in water bath (Taitec) with shaking (120 rpm) until luciferase activity is measured.
- Transfer 100 μl of culture to a well of a Black/Clear BD Falcon 96-well plate (Becton Dickinson) in triplicate for each culture.
Note: We tested 96-well plates for measuring luminescence in E. coli with the plate reader (Corona), indicating that the 96-well white plate showed the leakage of 1.5% to an adjacent empty well whereas the 96-well black plate reduced leakage by 0.95% (see the row data in both 96 plates in Figure 3). Although a white plate is usually used for luminescent measurements, a black plate was used to prevent leakage of high luminescence from one culture to other well in this study. Additionally, during measurement we arranged cultures with a single well gap between them to minimize leakage to adjacent wells. However, we recommend that a white plate be used first as standard procedure.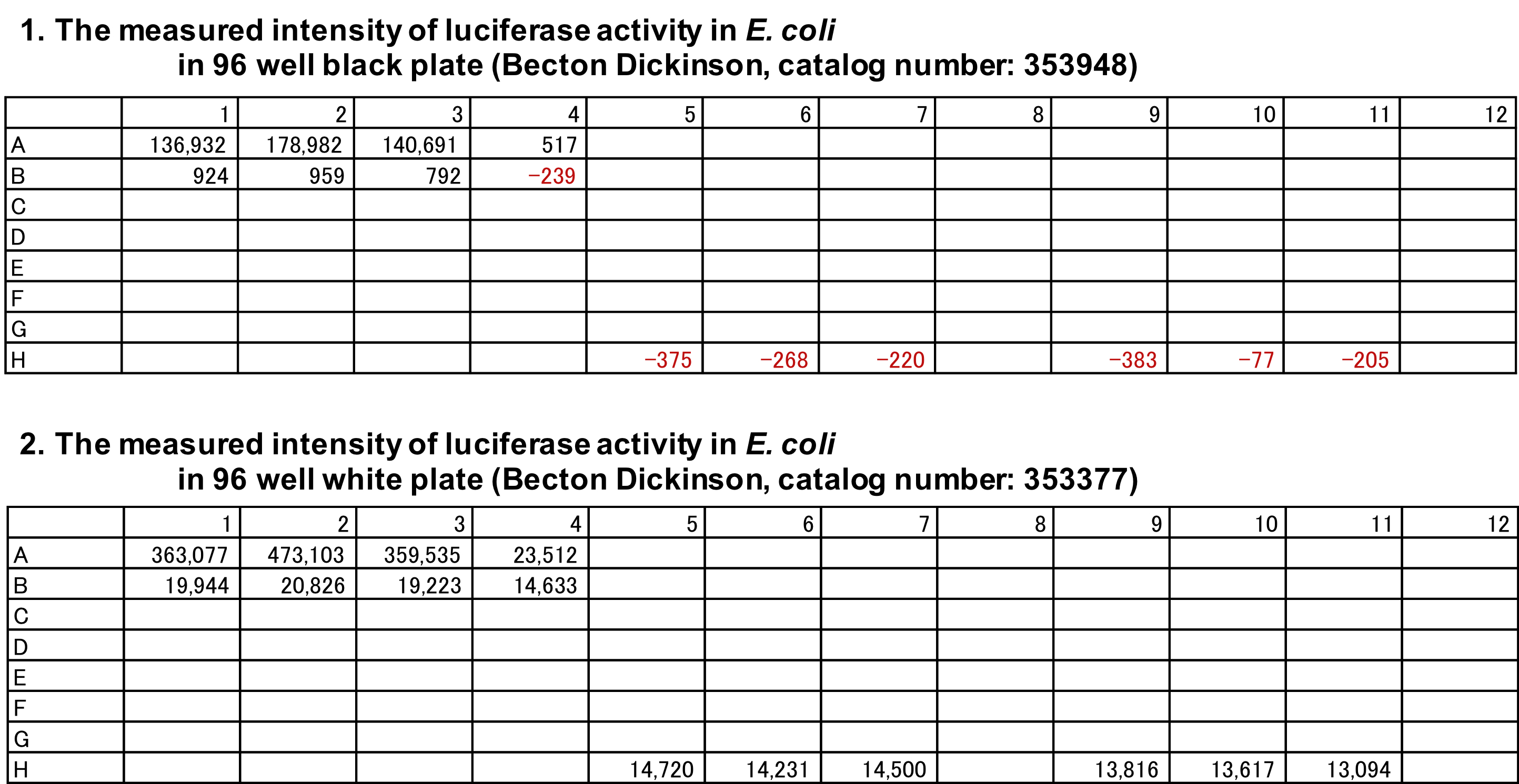
Figure 3. The measured intensity of luciferase activity in E. coli in 96-well plates. E. coli K-12 W3110 (parent strain) harbouring pLUXappYp, containing appY-lux operon (data not shown) was grown in M9-glucose medium at 37 °C, and culture was applied into three wells (A1, A2, and A3) of both the 96-well black plate (Becton Dickinson, catalog number: 353948) (upper) and the 96-well white plate (Becton Dickinson, catalog number: 353377) (lower). M9-glucose medium (H5, H6, and H7) and distilled water (H9, H10, and H11) were also applied into three wells of both the 96-well plates. Each intensity is shown as a raw data measured with the plate reader (Corona). The fluorescent intensity was detected in the empty wells adjacent to wells filled by culture (A4 and B1 to B4). - Transfer 100 μl of fresh M9-glucose medium to another well of the Black/Clear BD Falcon 96-well plate (Becton Dickinson) used in Step B6 in triplicate for background.
- Set the Black/Clear BD Falcon 96-well plate (Becton Dickinson) containing the samples and the background with plate reader (Corona).
- Measure OD600 according to the procedure for the plate reader (Corona).
- Measure luminescence as a total intensity. Therefore, no filter is set according to the procedure for the plate reader (Corona).
- Transform E. coli strains by the cloned luciferase reporter plasmids.
Data analysis
- Extract raw numeric data of both the values of OD600 and the intensities of luminescence as a text file from SF6 for Windows (Corona).
- Open a text file containing the raw numeric data in Microsoft Excel (Microsoft).
- Normalize the net values of OD600 and the net intensities of luminescence with background.
- Calculate the ratio of luminescence to OD600 as specific activity of the promoter of each culture by the following formula: the net intensity of luminescence/the net values of OD600.
- Average the ratios of triplicate with standard deviation.
Note: The activities of the 18 promoters used are represented in Figure 4, indicating that all 18 promoters silenced by H-NS were de-silenced by two H-NS mutants in agreement with our previous work (Yamanaka et al., 2018). To confirm a significant difference, calculated p-values by t-test of statistical analysis should be evaluated at less than 0.01.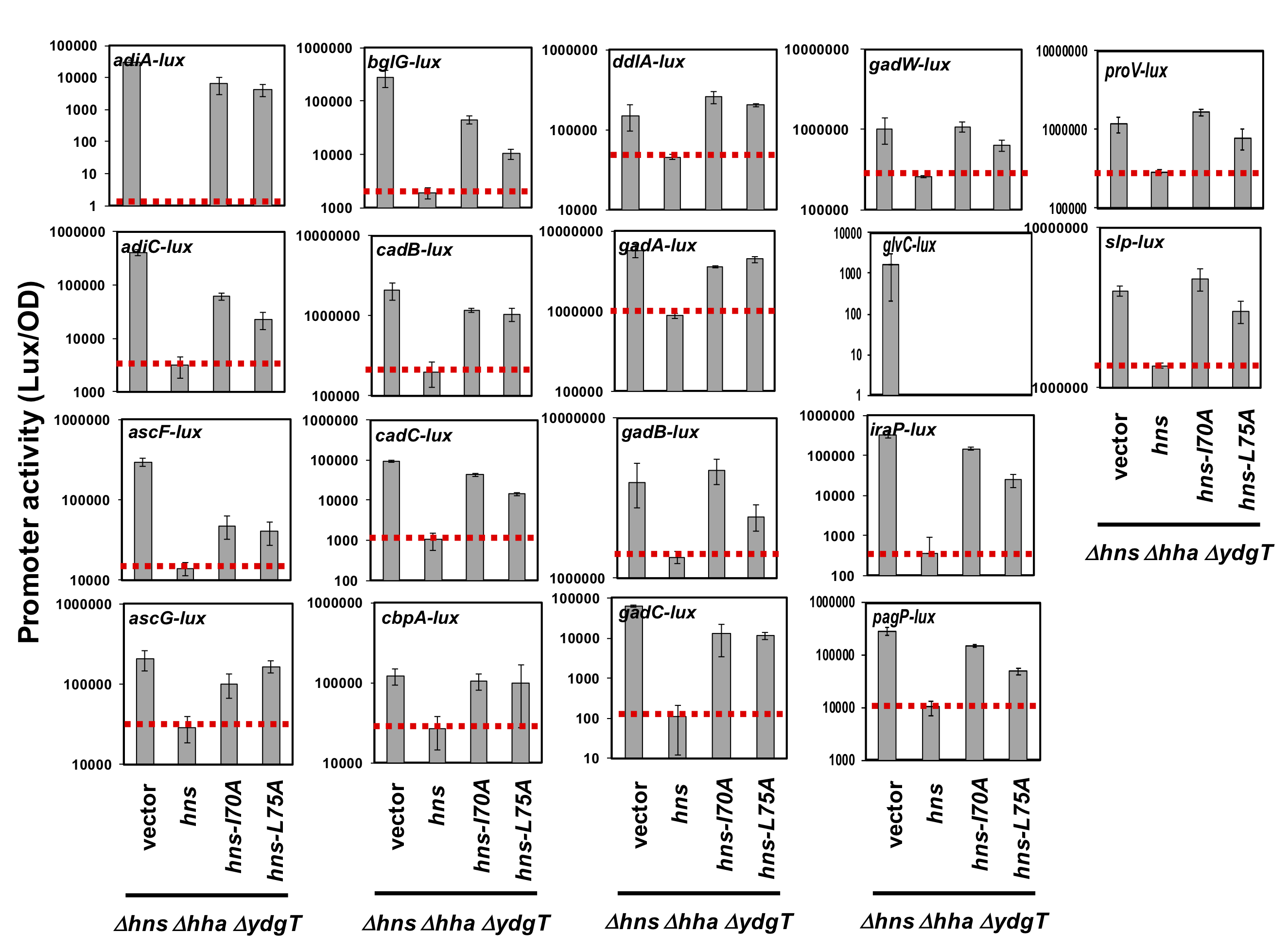
Figure 4. Luciferase reporter reveals silence of 18 promoters by H-NS in E. coli. The constructed 18 pLux reporter plasmids, pLUXadiAp, pLUXadiCp, pLUXascFp, pLUXascGp, pLUXbglGp, pLUXcadBp, pLUXcadCp, pLUXcbpAp, pLUXddlAp, pLUXgadAp, pLUXgadBp, pLUXgadCp, pLUXgadWp, pLUXglcVp, pLUXiraPp, pLUXpagPp, pLUXproVp, and pLUXslpp (Table 1), were introduced into ∆hns∆hha∆ydgT strains harbouring the hns plasmids (pQE80Lhns, pQE80Lhns-I70A, and pQE80Lhns-L75A) or an empty plasmid (a vector pQE80L). Transformants were grown in M9-glucose medium with 10 μM IPTG at 37 °C, and then the promoter activity was calculated as described above.
Recipes
- Competent E. coli DH5α
- Inoculate a single colony of E. coli DH5α in 5 ml of LB broth
- Incubate the pre-cultures at 37 °C with shaking for overnight
- Dilute overnight pre-culture 100-fold in 10 ml of fresh LB broth
- Incubate the cultures at 37 °C with shaking until mid-logarithmic phase
- Transfer 10 ml of culture to a sterile 50 ml plastic tube (Iwaki)
- Collect the cells by centrifugation (2,300 x g, 4 °C, 10 min)
- Suspend the cells in 10 ml of cold 0.1 M CaCl2
- Incubate the cells on ice for 30 min.
- Collect the cells by centrifugation (2,300 x g, 4 °C, 10 min)
- Suspend the cells in 1 ml of cold 0.1 M CaCl2
- Use a part (~100 μl) of the resuspended E. coli DH5α for transformation
- 50 mg/ml kanamycin
- Dissolve 0.5 g of kanamycin monosulfate (Nacalai Tesque) in 10 ml of distilled water
- Sterilize the solution by filtration with 0.22 μm filter (Advantec)
- Store the sterilized solution at 4 °C
- LB broth
- Dissolve 10 g of BactoTM tryptone, 5 g of BactoTM yeast extract, and 10 g of NaCl in 800 ml of distilled water
- Adjust pH to 7.5 with NaOH
- Adjust volume to 1 L with distilled water
- Autoclave the solution (set 121 °C and 20 min in LSX-500)
- Store the autoclaved LB broth at room temperature
- LB agar with 50 μg/ml kanamycin
- Dissolve 7.5 g of agar in 1 L of LB broth
- Autoclave (set 121°C and 20 min in LSX-500)
- Add kanamycin after cooling in the final concentration of 50 μg/ml
- Pour the media into Petri dishes, and then harden LB agar by cooling at room temperature
- Store the LB agar at 4 °C
- 125 mM EDTA
- Dilute 0.5 M EDTA (Nacalai Tesque) to 125 mM with a sterile water
- Store at room temperature
- 70% ethanol
- Dilute ethanol (Nacalai Tesque) to 70% with a sterile water
- Store at room temperature
- M9-Glucose medium (to autoclave, set 121 °C and 20 min in LSX-500)
- For 5x M9 salt (-NH4Cl2), dissolve 75 g of Na2HPO4∙12H2O, 15 g of KH2PO4 and 0.25 g of NaCl in 1 L distilled water, and then the dissolved solution was autoclaved
- Prepare the autoclaved following solutions: 2 M NH4Cl2, 1 M MgCl2 , 0.25 M K2SO4, 10 mM CaCl2
- Sterilize the 1 M glucose by filtration (0.22 μm filter)
- Add 200 ml of 5x M9 salt (-NH4Cl2), 10 ml of 2 M NH4Cl2, 1 ml of 1 M MgCl2 , 1 ml of 0.25 M K2SO4, 10 ml of 10 mM CaCl2 , 10 ml of 1 M Glucose into 768 ml of autoclaved distilled water
- Store the sterilized M9-glucouse medium at room temperature
Acknowledgments
We are extremely grateful to Prof. Akira Ishihama, Hosei Univeristy, for his comments and editing of the manuscript. We gratefully acknowledge Dr. Parisa Zangoui Nejad Chahkootahi (National University of Singapore) and Dr. Kyle David Buchan (National University of Singapore) for proofreading of the manuscript. The pLUX vector was a gift from Dr. Peter A. Lund, University of Birmingham. The E. coli ∆hns∆hha∆ydgT strain was a gift from Dr. Taku Oshima, Toyama Prefectural University. We thank Eri Arita, Kanako Hasegawa, Sho Watarai, Hideko Nakagawa, and Sato Fukunaga from Hosei university for technical assistance.
This protocol was adapted from Burton et al. (2010) for the method to luciferase reporter assay and modified from Yamanaka et al. (2018) for the method to high-throughput assay.
Competing interests
The authors declare no conflicts of interest.
References
- Bjarnason, J., Southward, C. M. and Surette, M. G. (2003). Genomic profiling of iron-responsive genes in Salmonella enterica serovar typhimurium by high-throughput screening of a random promoter library. J Bacteriol 185(16): 4973-4982.
- Burton, N. A., Johnson, M. D., Antczak, P., Robinson, A. and Lund, P. A. (2010). Novel aspects of the acid response network of E. coli K-12 are revealed by a study of transcriptional dynamics. J Mol Biol 401(5): 726-742.
- Meighen, E. A. (1991). Molecular biology of bacterial bioluminescence. Microbiol Rev 55(1): 123-142.
- Nozaki, S. and Niki, H. (2019). Exonuclease III (XthA) enforces in vivo DNA cloning of Escherichia coli to create cohesive ends. J Bacteriol 201(5): pii: e00660-18.
- Ueda, T., Takahashi, H., Uyar, E., Ishikawa, S., Ogasawara, N. and Oshima, T. (2013). Functions of the Hha and YdgT proteins in transcriptional silencing by the nucleoid proteins, H-NS and StpA, in Escherichia coli. DNA Res 20(3): 263-271.
- Yamanaka, Y., Oshima, T., Ishihama, A., and Yamamoto, K. (2014). Characterization of the YdeO regulon in Escherichia coli. PLoS ONE 9: e111962.
- Yamanaka, Y., Winardhi, R. S., Yamauchi, E., Nishiyama, S. I., Sowa, Y., Yan, J., Kawagishi, I., Ishihama, A. and Yamamoto, K. (2018). Dimerization site 2 of the bacterial DNA-binding protein H-NS is required for gene silencing and stiffened nucleoprotein filament formation. J Biol Chem 293(24): 9496-9505.
Article Information
Copyright
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Yamanaka, Y., Watanabe, H., Yamauchi, E., Miyake, Y. and Yamamoto, K. (2020). Measurement of the Promoter Activity in Escherichia coli by Using a Luciferase Reporter. Bio-protocol 10(2): e3500. DOI: 10.21769/BioProtoc.3500.
- Yamanaka, Y., Winardhi, R. S., Yamauchi, E., Nishiyama, S. I., Sowa, Y., Yan, J., Kawagishi, I., Ishihama, A. and Yamamoto, K. (2018). Dimerization site 2 of the bacterial DNA-binding protein H-NS is required for gene silencing and stiffened nucleoprotein filament formation. J Biol Chem 293(24): 9496-9505.
Category
Microbiology > Microbial genetics > Gene expression
Molecular Biology > DNA > Gene expression
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link