- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Bone-in-culture Array to Model Bone Metastasis in ex vivo Condition
Published: Vol 10, Iss 2, Jan 20, 2020 DOI: 10.21769/BioProtoc.3495 Views: 4965
Reviewed by: HongLok LungDipak Kumar PoriaAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

3D Stroma Invasion Assay
Yvette May Coulson-Thomas and Vivien Jane Coulson-Thomas
Mar 20, 2017 9331 Views

Calvarial Bone Implantation and in vivo Imaging of Tumor Cells in Mice
Kyoko Hashimoto [...] Mitsuru Futakuchi
Feb 5, 2019 8173 Views

Cell-derived Matrix Assays to Assess Extracellular Matrix Architecture and Track Cell Movement
Kendelle J. Murphy [...] David Herrmann
Dec 20, 2022 3354 Views
Abstract
Bone is the most frequently affected organ by metastases of breast cancer and prostate cancer. Our knowledge on bone metastasis is extremely limited due to the lack of potent and efficient experimental models. We developed the “Bone-In-Culture Array (BICA)” platform to model the bone colonization of cancer cells in ex vivo cultures. The use of the BICA platform will facilitate in-depth mechanistic studies and high-throughput screening of drugs for bone metastasis.
Keywords: Bone metastasesBackground
Bone metastasis research is extremely limited due to the lack of efficient and faithful experimental models (Weigelt et al., 2005; Simmons et al., 2015). Current in vivo bone metastasis models require extensive surgical labor, rendering high-throughput assays impossible (Blouin et al., 2005; Simmons et al., 2015; Kuchimaru et al., 2018). In vitro systems, on the other hand, cannot recapitulate the cellular and molecular complexity of the bone microenvironment (Taubenberger, 2014; Cunningham and You, 2015). To complement existing systems, we developed the BICA platform to investigate mechanisms involved in bone colonization and to rapidly test drug efficacies on bone metastases (Wang et al., 2015; Yu et al., 2016).
BICA is performed by selectively delivering cancer cells into the hind limbs of mice via the external iliac artery followed by extracting and fragmenting the cancer-containing bones and exposing the bone fragments to ex vivo cultures (Wang et al., 2015; Yu et al., 2016). The cancer cell growth in BICA will then be determined using bioluminescence signals. Our previous work has established that BICA recapitulates in vivo bone metastases characteristics, including the microenvironmental niche, gene expression profile, metastatic growth kinetics, and therapeutic responses (Wang et al., 2015). Therefore, BICA as a reliable and high-throughput experimental platform to investigate bone metastases processes in breast cancer and prostate cancer or even other cancer types that can potentially give rise to bone metastases.
Materials and Reagents
- 31 G needles (BD, catalog number: 08290-3284-18)
- 0.45 µm polyethersulfone membranes (VWR, catalog number: 28145-505)
- BICA plate: 96-well low attachment plate (Corning, catalog number: 3474)
- Bone plier (F.S.T., catalog number: 16025-14)
- pwpt-GFP (Addgene, catalog number: 12255)
- pMD2.G (Addgene, catalog number: 12259)
- psPAX2 (Addgene, catalog number: 12260 )
- Fetal Bovine Serum, qualified, United States (Gibco, catalog number: 26140079)
- DMEM/F-12 with GlutaMAXTM supplement1 (Gibco, catalog number: 10565018)
- Ethanol (KOPTEC, catalog number: 64-17-5)
- Trypsinize (with Trypsin 0.25% Solution; HyClone, catalog number: SH30042.01)
- Saline (Teknova, catalog number: S5815)
- 10x luciferase substrate D-Luciferin (Goldbio, catalog number: LUCNA-1G)
- Culture media (see Recipes)
Equipment
- Micro-dissecting scissor (Roboz, catalog number: RS-5912)
- Micro-dissecting forceps (Roboz, catalog number: RS-5135)
- IVIS Lumina II in vivo Imaging System (PerkinElmer)
- Dissecting microscope (Leica S6E stereomicroscopes with KL200 LED light sources)
- CO2 Incubator (Thermo Scientific, Forma Series II water Jacket)
- FACS
Software
- Living Image Software-IVIS Lumina Series (PerkinElmer, v4.2)
Procedure
- Intrailliac artery injection (IIA)
- Cell preparation
Depending on research purposes, IIA injection can be performed with different cancer cell lines of variable origins and cancer types. We typically use Firefly luciferase tagged cancer cells, whose progression can be monitored by bioluminescence signal. The preparation of luciferase-tagged cancer cells was described in our previous study (Wang et al., 2017). Briefly, replace the GFP domain of pwpt-GFP (Addgene #12255) with Fluc-GFP fusion gene. Transfect 293T cells with 5 µg pwpt-Fluc-GFP vector, along with 1.75 µg pMD2.G (Addgene #12259) and 3 µg psPAX2 (Addgene #12260) in a 10 cm cell culture dish to package lentivirus. After 48 h, filter the supernatant through 0.45 µm polyethersulfone membranes (VWR), which will serve as lentiviral stocks. Treat the cancer cells with Fluc-GFP lentivirus and 4 mg/ml polybrene for 8 h. After a 72 h culture period, isolate the GFP-positive cells by FACS. Cells are maintained by following their corresponding culture protocols.- Upon harvesting, trypsinize (with Trypsin 0.25% Solution), wash and resuspend cancer cells to a concentration of 5 x 106 cells/ml in ice-cold, sterilized saline (Teknova).
- Keep cell suspensions on ice.
- Mice preparation
Choose mice in the appropriate gender and ages based on the purpose of the conducted research. Of note, the immune rejection between different species (e.g., human and mice) or stains will not be preserved in ex vivo culture. Therefore, researcher may have more permissive choice in determining the mice strains to use.
The anesthesia and aseptic procedure should follow the standard operation procedure approved by IACUC in the researcher’s institute. - IIA injection procedure
In this step, we selectively inject cancer cells via the external iliac artery and thereby loading the cancer cells into the hind limb of mice. Here we briefly introduce the major steps of IIA as the basic work for BICA samples preparation. The details of this procedure are elaborately described in our previous publications (Wang et al., 2015; Yu et al., 2016), which is out of the main scope of this study.- Restrain the anesthetized mice on a diaper or plastic board.
- Make a skin incision of 1.5 cm between the 4th and 5th nipples in the lower right abdomen.
- Perform blunt dissection to separate muscles and expose the common iliac artery under the dissection microscope.
- Inject 0.1 ml cell suspension into the iliac artery using 31 G needles.
- Stop bleeding by pressing injected artery with cotton tips (in about 10 min). This step may take 5-10 min.
- Leave the mouse on a warm heating pad under anesthesia and aseptic condition for 30 min, allowing the cancer cells homing to the favorable niches.
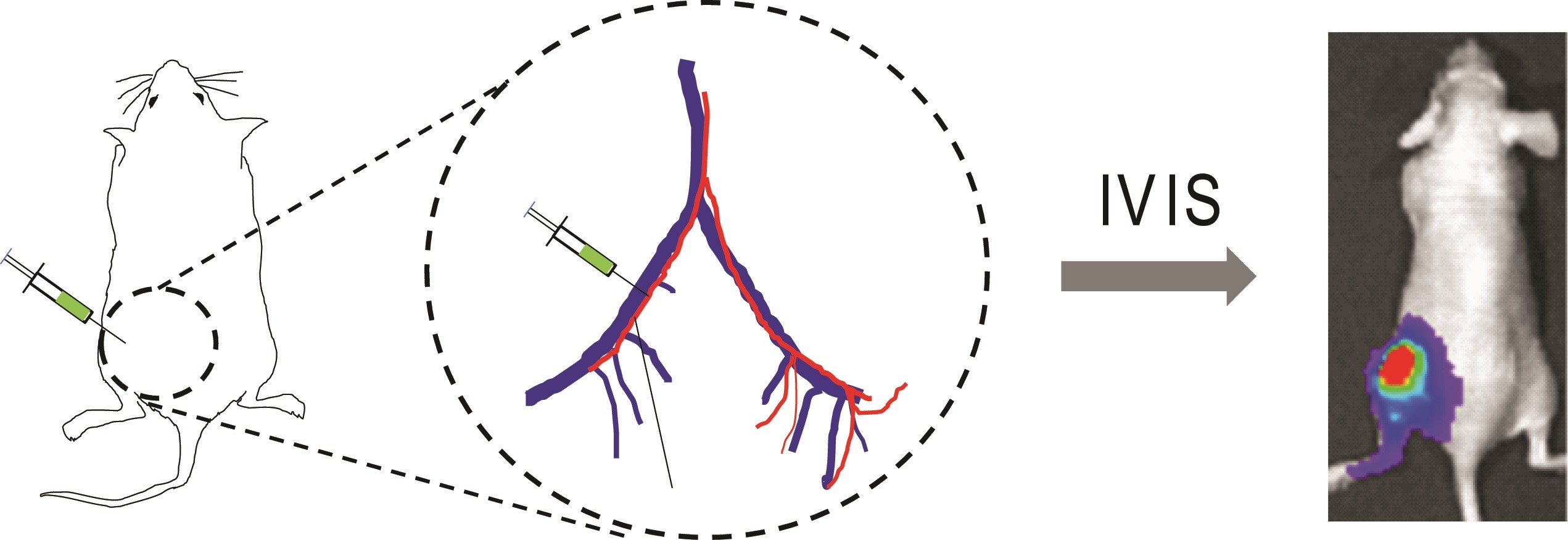
Figure 1. Schematic of IIA injection. Luciferase-tagged cancer cells are inoculated into the external iliac artery of mouse, which can be tracked by bioluminescence imaging.
- Cell preparation
- Preparation of BICA samples
In order to reduce the chance of microbio-contamination, this step should be performed within the aseptic environment such as a laminar flow cabinet.- Bone collection
- Thirty minutes after receiving intra-iliac injection, euthanize the mice with the standard operation procedure approved by IACUC in the researcher’s institute.
- Spray the mice body, especially the leg receiving IIA injection with 70% ethanol for sterilization.
- Extend the incision made for IIA injection, and stripe the skin of the hind limb.
- Remove the muscles and tendons carefully with scissor.
- Wipe bone with sterilized gauzes to clear the residual non-bone tissues, thereby fully exposing the femur and tibia bones.
- Bone trimming
- In a sterilized 6 cm cell culture dish, cut down the distal epiphysis and metaphysis of the femurs, and the proximal epiphysis and metaphysis of the tibias. These parts are bone tissues mostly enriching colonized cancer cells.
- Crush the bone tissues with a sterilized bone plier (F.S.T.), trim the bone fragments with dissecting scissor (Roboz) to an even size (0.5-1.5 mm in diameter).
Note: Due to the tight interaction already formed between cancer cell and bone matrix, cancer cells are confined within bone fragments during this time. However, do not leave the samples exposed in the air for longer than 5 min. - Quickly transfer the bone fragments to low-attachment 96-well plates preloaded with 200 µl culture media (one piece per well). If necessary, randomize bone fragments from multiple mice in different group settings.
- Bone collection
- (Optional) Observation of the baseline intensity by bioluminescence imaging
Researchers in lack of experience may create large deviation on fragment size and cancer loads correspondingly. In this scenario, the bioluminescence intensity data observed later can be divided over the Day 0 (or Day 1) signal intensity of the same piece to yield normalized values, which will help to reduce the intragroup deviation.- Bioluminescence imaging
- Use multi-pipette to add 10x luciferase substrate D-Luciferin into sample wells.
- Image the plate with an IVIS Lumina II machine. Field of view = D; bining = Medium; F/Stop = Exposure time could be adjusted (increased or reduced) upon the bioluminescence intensity of the cancer cells.
- Replace the media containing D-luciferin
The residual substrate may exhaust the luciferase expressed by cancer cells, therefore interfere in the future measurements. Therefore, every time after imaging, we need to change the media to remove the residual substrate.- Use multi-pipette to remove the medium along the wall of wells. Avoid fragment aspiration.
- Rinse each well with 200 µl sterilized PBS.
- Use multi-pipette to remove the PBS along the wall of wells. Avoid fragment aspiration.
- Add 200 µl fresh culture media into each well.
- Bioluminescence imaging
- Ex vivo culturing and imaging
The duration of culturing and imaging frequency depends on the characteristics of the cells loaded in BICA samples. Usually, murine cancer cells progress faster than human cells; Breast cancer cells progress faster than prostate cancer cells; both of which are in agreement with their growth kinetics in vivo.- BICA Culture
- Culture the BICA samples in a 37 °C incubator with 5% CO2 supply. In studies monitoring cancer biology in low oxygen conditions, a hypoxia chamber can be used in place of the cell culture incubator.
- Change the media every 3-4 days, or when media turns orange-yellow.
- Measure the BICA plate at least once a week by bioluminescence imaging.
- The traits of bone residual cells can be maintained for as long as 2 months.
- Bioluminescence imaging
The bone-colonized cancer cells are usually tightly anchored into the bone matrix. Floating cancer cells are very rare, which won’t interrupt the measurement of bioluminescence intensity.- Use multi-pipette to add 10x luciferase substrate D-Luciferin into sample wells.
- Image the plate with IVIS Lumina II machine. Field of view = D; bining = Medium; F/Stop = 1. Exposure time could be adjusted (increased or reduced) upon the bioluminescence intensity of the cancer cells.
- Replace the media containing D-luciferin:
- Use multi-pipette to remove the medium along the wall of wells. Avoid fragment aspiration.
- Rinse each well with 200 µl sterilized PBS.
- Use multi-pipette to remove the PBS along the wall of wells. Avoid fragment aspiration.
- Add 200 µl fresh culture media into each well.
- Immunohistochemistry (IHC) and immunofluorescence (IF) imaging
The IHC/IF staining is one of the extensional applications of BICA. If needed, fix the BICA tissues with 4% Paraformaldehyde overnight and decalcify them with 14% EDTA for five days. Paraffin-embed the samples for micro-sectioning. ALP and CTSK can serve as markers for osteoblast and osteoclast individually. More details can be found in our previous studies (Wang et al., 2015, 2017 and 2018).
- BICA Culture
Data analysis
The bioluminescence intensity data observed later can be interpreted by either raw or normalized intensities. In the latter case scenario, divide the bioluminescence intensity by the signal intensity of the same piece on the Day 0 (or Day 1), which may help to reduce the intragroup deviation (Figure 2).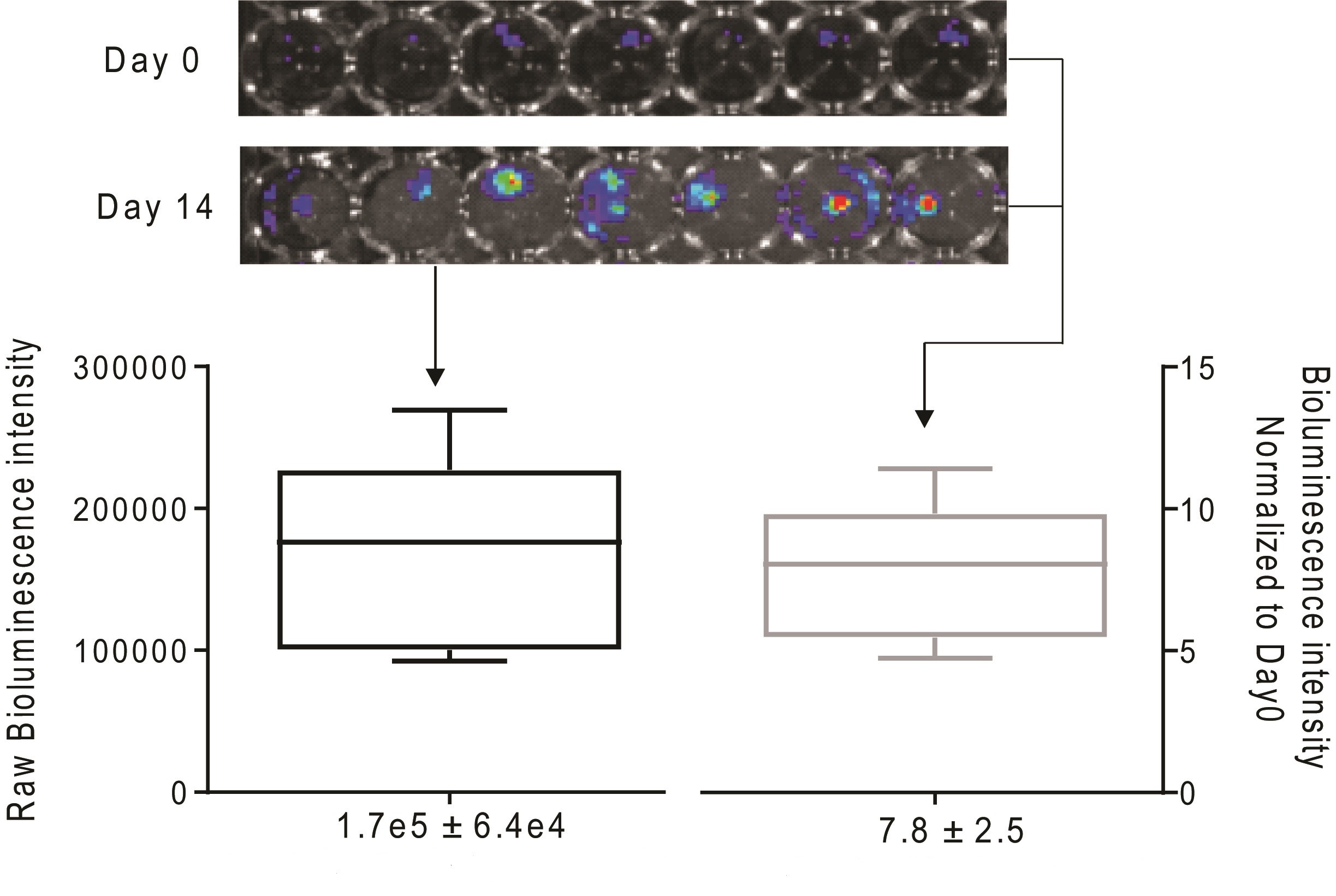
Figure 2. Quantification of bioluminescence signal derived from BICA samples of prostate cancer cell line 22rv1 by either raw reads (left) or normalized intensities (right). SD = 0.38 fold of mean value (left). SD = 0.32 fold of mean value (right).
Recipes
- Culture media
BICA samples with human cancer cell lines: DMEM/F12 + 2% FBS
BICA samples with mouse cancer cell lines: DMEM/F12 + 0.5% FBS
Acknowledgments
We thank Zhang Laboratory members for the helpful input. X.H.-F.Z. is supported by the US Department of Defense DAMD W81XWH-16-1-0073, NCI CA183878, the Breast Cancer Research Foundation, and the McNair Medical Institute. H.W. is supported by the US Department of Defense DAMD W81XWH-13-1-0296.
The protocol is a detailed technical summary of: Wang et al. (2018) Cancer Cell 34(5): 823-839 and Wang et al. (2017) Nat Commun 8: 15045.
Competing interests
The author declares no competing financial interests.
Ethics
All animal work was done in accordance to the animal care guidelines of the Baylor College of Medicine.
References
- Blouin, S., Baslé, M. F. and Chappard, D. (2005). Rat models of bone metastases. Clin Exp Metastasis 22(8): 605-614.
- Cunningham, D. and You, Z. (2015). In vitro and in vivo model systems used in prostate cancer research. J Biol Methods 2(1).
- Kuchimaru, T., Kataoka, N., Nakagawa, K., Isozaki, T., Miyabara, H., Minegishi, M., Kadonosono, T. and Kizaka-Kondoh, S. (2018). A reliable murine model of bone metastasis by injecting cancer cells through caudal arteries. Nat Commun 9(1): 2981.
- Simmons, J. K., Hildreth, B. E., 3rd, Supsavhad, W., Elshafae, S. M., Hassan, B. B., Dirksen, W. P., Toribio, R. E. and Rosol, T. J. (2015). Animal models of bone metastasis. Vet Pathol 52(5): 827-841.
- Taubenberger, A. V. (2014). In vitro microenvironments to study breast cancer bone colonisation. Adv Drug Deliv Rev 79-80: 135-144.
- Wang, H., Tian, L., Goldstein, A., Liu, J., Lo, H. C., Sheng, K., Welte, T., Wong, S. T. C., Gugala, Z., Stossi, F., Zong, C., Li, Z., Mancini, M. A. and Zhang, X. H. (2017). Bone-in-culture array as a platform to model early-stage bone metastases and discover anti-metastasis therapies. Nat Commun 8: 15045.
- Wang, H., Tian, L., Liu, J., Goldstein, A., Bado, I., Zhang, W., Arenkiel, B. R., Li, Z., Yang, M., Du, S., Zhao, H., Rowley, D. R., Wong, S. T. C., Gugala, Z. and Zhang, X. H. (2018). The osteogenic niche is a calcium reservoir of bone micrometastases and confers unexpected therapeutic vulnerability. Cancer Cell 34(5): 823-839 e7.
- Wang, H., Yu, C., Gao, X., Welte, T., Muscarella, A. M., Tian, L., Zhao, H., Zhao, Z., Du, S., Tao, J., Lee, B., Westbrook, T. F., Wong, S. T., Jin, X., Rosen, J. M., Osborne, C. K. and Zhang, X. H. (2015). The osteogenic niche promotes early-stage bone colonization of disseminated breast cancer cells. Cancer Cell 27(2): 193-210.
- Weigelt, B., Peterse, J. L. and van 't Veer, L. J. (2005). Breast cancer metastasis: markers and models. Nat Rev Cancer 5(8): 591-602.
- Yu, C., Wang, H., Muscarella, A., Goldstein, A., Zeng, H. C., Bae, Y., Lee, B. H. and Zhang, X. H. (2016). Intra-iliac artery injection for efficient and selective modeling of microscopic bone metastasis. J Vis Exp (115).
Article Information
Copyright
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Wang, H. and Zhang, X. H. (2020). Bone-in-culture Array to Model Bone Metastasis in ex vivo Condition. Bio-protocol 10(2): e3495. DOI: 10.21769/BioProtoc.3495.
Category
Cancer Biology > Invasion & metastasis > Tumor microenvironment
Cell Biology > Tissue analysis
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









