- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Measurement of Chloroplastic NAD Kinase Activity and Whole Tissue NAD Kinase Assay
(*contributed equally to this work) Published: Vol 10, Iss 1, Jan 5, 2020 DOI: 10.21769/BioProtoc.3480 Views: 4878
Reviewed by: Zhibing LaiGuotian LiAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Seed Collection in Temperate Trees—Clean, Fast, and Effective Extraction of Populus Seeds for Laboratory Use and Long-term Storage
Naima Bhutta [...] Katharina Bräutigam
Feb 5, 2024 2036 Views

Vegetative Propagation of Cannabis sativa and Resin Obtained From its Female Inflorescences
Sebastián D´Ippolito [...] Silvana L. Colman
Feb 20, 2025 1735 Views

A New Approach to Detect and Semi-quantify All Molecular Species and Classes of Anionic Phospholipids Simultaneously in Plant Samples
Manon Genva [...] Laetitia Fouillen
Apr 20, 2025 1718 Views
Abstract
Nicotinamide adenine dinucleotide phosphate (NADP) synthesis requires nicotinamide adenine dinucleotide (NAD) kinase activity, substrate NAD and ATP. The NAD kinase responds to various environmental stimuli and its activity is regulated via various regulatory pathways, such as Ca2+-dependent and redox-dependent signals. Conventional in vitro NAD kinase assay has been useful to evaluate enzyme activity; however, recent reports revealed a dynamics of NADP pool (the sum of NADP+ and NADPH) under fluctuating light condition, indicating that the rate of NADP synthesis is not always determined by NAD kinase activity. Here, we developed a novel method for the estimation of chloroplastic NAD kinase activity by quantifying the changes in the NADP amounts in response to illumination. As our approach does not involve protein extraction, it saves time (compared to the in vitro assay), thereby allowing for a sequence of assays, and provides several clues in the investigation of regulatory mechanisms behind NADP synthesis under various environmental conditions.
Keywords: NAD kinaseBackground
NADP (including the oxidized form NADP+ and the reduced form NADPH) is known as one of the most fundamental molecules for cellular metabolic reactions. In nature, NADP+ captures light-driven energy as electrons, via the photosynthetic electron transfer chain (PETC). Therefore, the level of available NADP+ in photosynthesis is quite important for autotrophic growth (Hashida and Kawai-Yamada, 2019).
Plant NAD kinase activity has been evaluated in many reports. According to in vitro assay on Arabidopsis thaliana tissue extracts, whole NAD kinase activity in 2-week-old seedlings was estimated as 560 nmol/h/mg protein (Turner et al., 2004). On the other hand, in 3-week-old rosette leaves of Col-0 it was calculated as 5.3 nmol/h/mg protein and 64 nmol/h/g FW (corresponding to approximately 12.1 nmol/h/mg protein) (Takahashi et al., 2006 and 2009). This large difference might be from variations in the amount of RubisCO (the most abundant leaf protein) between juvenile and mature leaves. Furthermore, it has been known that NAD kinase activity is under the control of various environmental stimuli, such as light, pathogen infection, oxidative stress etc. (Berrin et al., 2005). Beside transcriptional responses, several plant NAD kinases are known to be regulated via Ca2+ and calmodulin (CaM) signaling, cellular redox states and phytochromes (Tezuka and Yamamoto,1975; Delumeau et al., 2000; Hashida et al., 2018; Dell’ Aglio et al., 2019; Tai et al., 2019). Therefore, robust, reproducible, and standardized methods are indispensable for the evaluation of NAD kinase activity.
In Arabidopsis, the nadk2, a loss-of-function mutant in the chloroplastic NAD kinase (At1g21640) (Chai et al., 2005), lacked the light-responsive increase in NAD kinase activity (Hashida et al., 2018), suggesting that the activity of the NADK2 isoform is highly dependent on the light condition. Here we developed a novel NAD kinase assay, especially focusing on the chloroplastic NAD kinase activity. Compared to conventional in vitro NAD kinase assay, our newly developed method can drastically save time and costs, and it enables the evaluation of chloroplastic NAD kinase activity, while being comparable to the conventional method.
Materials and Reagents
- Finntip Pipette Tips (Finntip Flex 10, 200, and 1,000)
- 1.5 ml microcentrifuge tubes (VIOLAMO, F 1.5, catalog number: 1-1600-01)
- 12 Well Multidish (Thermo Fisher Scientific, catalog number: 130185)
- Half area 96-Well microplate (Greiner Bio-One, catalog number: 675075)
- ZebaTM Spin Desalting Columns, 40K MWCO, 0.5 ml (Pieces, catalog number: 87766)
- Arabidopsis thaliana wild-type plants (Ecotype: Col-0)
- Liquid nitrogen
- Tris (hydroxymethyl) aminomethane (HEPES) (Nacalai Tesque, catalog number: GR35406-91)
- MgCl2 (Nacalai Tesque, catalog number: SP20937)
- CaCl2 (Nacalai Tesque, catalog number: SP08894)
- PMSF (Nacalai Tesque, catalog number: SP96297)
- cOmpleteTM, Mini, Protease Inhibitor Cocktail (Roche, catalog number: 4693159001)
- Bio-Rad Protein Assay Kit (Bio-Rad, catalog number: 5000001ja)
- NADP/NADPH-GloTM Assay Kit (Promega, catalog number: G9081 or G9082)
- NaH2PO4 (Nacalai Tesque, catalog number: SP31737)
- ATP (Nacalai Tesque, catalog number: SP08886)
- NAD+ (Nacalai Tesque, catalog number: GR24338)
- NADP+ (Nacalai Tesque, catalog number: EP24336)
- NADPH (Nacalai Tesque, catalog number: CP24340)
- HCl (Nacalai Tesque, catalog number: GR18322)
- NaOH (Nacalai Tesque, catalog number: SP06338)
- Protein extraction buffer (see Recipes)
- Reaction buffer (see Recipes)
- Luciferin detection reagent (see Recipes)
- NADP/NADPH Glo detection reagent (see Recipes)
Equipment
- Pipettes (Thermo Fisher Scientific, FinnpipetteTM F2)
- Conventional cork borer (4.0 mm diameter)
- Homogenizer pestle (VIOLAMO, F-1.5, catalog number: 1-2955-02)
- Refrigerated microcentrifuge (TOMY, model: MX305)
- Ultrasonic disruptor (TOMY, model: UR-21P)
- Absorption spectrometer (Thermo Fisher Scientific, MultiskanTM GO)
- Luminescence detector (Promega, model: GloMaxTM Navigator System GM2000)
Procedure
- Setting up a conventional in vitro NAD kinase assay
- Protein extraction
- Grind freshly detached leaves (200-400 mg) in liquid nitrogen with a mortar and pestle, and add 300 µl of the ice-cold protein extraction buffer (prepared immediately before use, Recipe 1).
Note: Our laboratory normally uses three to four week-old rosette leaves, sampled midday. As NAD(P) metabolism could dynamically change during the phase transition from vegetative to reproductive growth (Hashida et al., 2016), be sure to prepare samples before bolting. We recommend at least three biological replicates and two technical replicates to obtain reliable results. - Further disrupt leaf sample by ultrasonic processing for 10 s with output control 6 on ice. Repeat this step two times to thoroughly rupture chloroplasts.
Note: According to manufacturer’s specifications of the equipment (UR-21P), output control 6 responds to 6 Watts. Ultrasonic frequency is 28 kHz and exponential horn has 2.5 mm diameter. - After centrifugation at 14,000 x g, 4 °C for 10 min, transfer supernatant (i.e., crude extract) to a 1.5 ml microcentrifuge tube to remove sample debris.
- Remove NAD+, NADH, NADP+ and NADPH using ZebaTM Spin Desalting Columns by a 4 °C centrifugation at 14,000 x g for 2 min and collect the flow-through containing the extracted proteins in a new 1.5 ml microcentrifuge tube.
- Grind freshly detached leaves (200-400 mg) in liquid nitrogen with a mortar and pestle, and add 300 µl of the ice-cold protein extraction buffer (prepared immediately before use, Recipe 1).
- Protein measurement
- Prepare the dye for the Bio-Rad Protein Assay by diluting 1 volume of the dye concentrate with 4 volumes of distilled, deionized (DDI) water.
- Add 2 and 5 µl of protein extracts into 198 and 195 µl diluted dye reagent, respectively.
- After 5 min, measure absorbance at 595 nm and estimate the protein concentration with the protein standard II (BSA) from the Bio-Rad Protein Assay Kit (0.2 to 2.0 mg/ml).
- NAD+ phosphorylation reaction
- Mix the ingredients in a new 1.5 ml tube bellow in the following amounts:
Extracted proteins, 10 µg
Reaction buffer (Recipe 2), 10 µl
50 mM NAD+, 10 µl
50 mM ATP, 10 µl - Incubate the mixture at 30 °C for 30 min in a water bath.
- Terminate reaction by heating the sample at 95 °C for 2 min.
- After centrifugation at 14,000 x g, 4 °C for 5 min, use the supernatant for NADP measurement (designated here as “Measuring solution”).
- Mix the ingredients in a new 1.5 ml tube bellow in the following amounts:
- Protein extraction
- Setting up the chloroplastic NAD kinase assay
- NAD+ phosphorylation reaction
- Float freshly prepared leaf discs in a 12-well plate on DDI water, and place the plate under dark for 1 h (dark acclimation).
Note: Our laboratory uses only 12 leaf discs (with 4 mm diameter) per well to avoid cross-shading during light exposure (Figure 1). - Transfer the plate under light (80 µmol photons/(s·m2)) and start a time-course for the termination of the reaction, by collecting two discs into a 1.5 ml microcentrifuge tube containing 80 µl of 0.2 N HCl (for the quantification of the oxidized form, NADP+), and another two discs into 80 µl of 0.2 N NaOH (for the quantification of the reduced form, NADPH).
Note: Be sure to prepare blank controls (no leaf discs in the tube). - Heat the sample for 2 min at 95 °C in a hot water bath, and thoroughly ram the discs by pestle. After adding 8 µl of 200 mM sodium-phosphate solution (pH 5.6), adjust pH between 5 and 6 with the appropriate volumes of 0.2 N NaOH or 0.2 N HCl.
Note: Check the pH with a test paper. This pH adjustment usually requires 50 to 60 µl NaOH and HCl solution.
- Float freshly prepared leaf discs in a 12-well plate on DDI water, and place the plate under dark for 1 h (dark acclimation).
- After centrifugation at 14,000 x g, 4 °C for 5 min, use the supernatant for NADP measurement, designated here as the “Measuring solution”.

Figure 1. Leaf discs for the chloroplastic NAD kinase assay
- NAD+ phosphorylation reaction
- NADP measurement
- Preparing detection reagent for the NADP/NADPH-GloTM assay
- For luminescence-based NADP detection, prepare the required amount of luciferin detection reagent (Recipe 3) before all the assays, and stored it at room temperature (approximately 23 to 25 °C). When you obtain the “Measuring solution” (see above), prepare the NADP/NADPH-Glo detection reagent just before use (Recipe 4).
- For the calibration curve, prepare the standard using 1 µM NADP+ and 1 µM NADPH. Add 0, 1, 2, 3, 4 and 5 µl of NADP+ or NADPH standard into a series of wells in a half area 96-well microplate and adjust the volume to 50 µl per well with each blank control.
- Transfer 50 µl of the measuring solution, which was obtained in the Steps A3d or B1d, into the other empty wells.
- Luminescence-based detection with NADP/NADPH-GloTM assay
- Add 50 µl of the NADP/NADPH-Glo detection reagent to each well, and briefly shake plates on a microplate shaker (gently). Incubate for 20 min at room temperature (approximately 23 to 25 °C) in the dark.
- Detect and record luminescence using a GloMax Navigator System. Set the integration at 0.3 s per well.
Note: Repeat the detection after 5 to 10 min and check whether the luminescence count was saturated or not. If the count was saturated, dilute the measuring solution and repeat the detection procedure to avoid underestimation.
- Preparing detection reagent for the NADP/NADPH-GloTM assay
Data analysis
- Calculation of the NADP+ and NADPH quantity from the luminescence units (U.I.) needs to be done by using the standard curve. Values acquired from this protocol will represent the quantity of NADP+ and NADPH molecules in the measuring solution. In the in vitro assay, NADPH could be sometimes detected even if NAD+ was used for the unique substrate, due to contaminating activity from the NADP+ reduction. In order to avoid underestimation, for the calculation of activity we recommend summing up the NADP+ and NADPH quantities as nmol NADP/h/mg protein or as nmol NADP/h/g fresh weight (FW) as follows:
[sum of NADP+ and NADPH (nmol)] × [volume of the measuring solution (µl)]/[used volume for detection (50 µl)]/[reaction time (h)]/[protein amount (mg) or sample weight (g fresh weight)] - Unlike for the in vitro assay, NADPH quantity is required to estimate NAD+ kinase activity by the approach with leaf disc, simply because PETC generates NADPH from NADP+ during illumination. A representative result is shown in Figure 2. Increase in the NADP, which is shown as a sum of NADP+ and NADPH, is never observed under complete dark condition (Figure 2A, Dark). Here, calculation of the NADP+ and NADPH quantity for each time points was done as follows:
[sum of NADP+ and NADPH (nmol)] × [volume of measuring solution (µl)]/[used volume for detection (50 µl)]/[leaf area (cm2)] - NADP content can also be expressed as nmol NADP/mg Chlorophyll (Chl) or nmol NADP/h/g FW, for which the appropriate measures (quantification of the total Chl content, or measuring the weight of the leaf discs) would need to be taken, before the assay.
- Light-driven NADP increase can usually be detected within 30 s. In the first approx. 10 min, NADP increase under illumination was proportional to time, as shown in Figure 2A. Figure 2B illustrates the increase in NADP at the beginning of illumination, as nmol NADP/g FW. Hence, the chloroplastic NAD kinase activity could be calculated as follows:
[sum of NADP+ and NADPH (nmol)] × [volume of measuring solution (µl)] × [60 (min)]/[used volume for detection (50 µl)]/[time after light exposure (min)]/[sample weight (g)]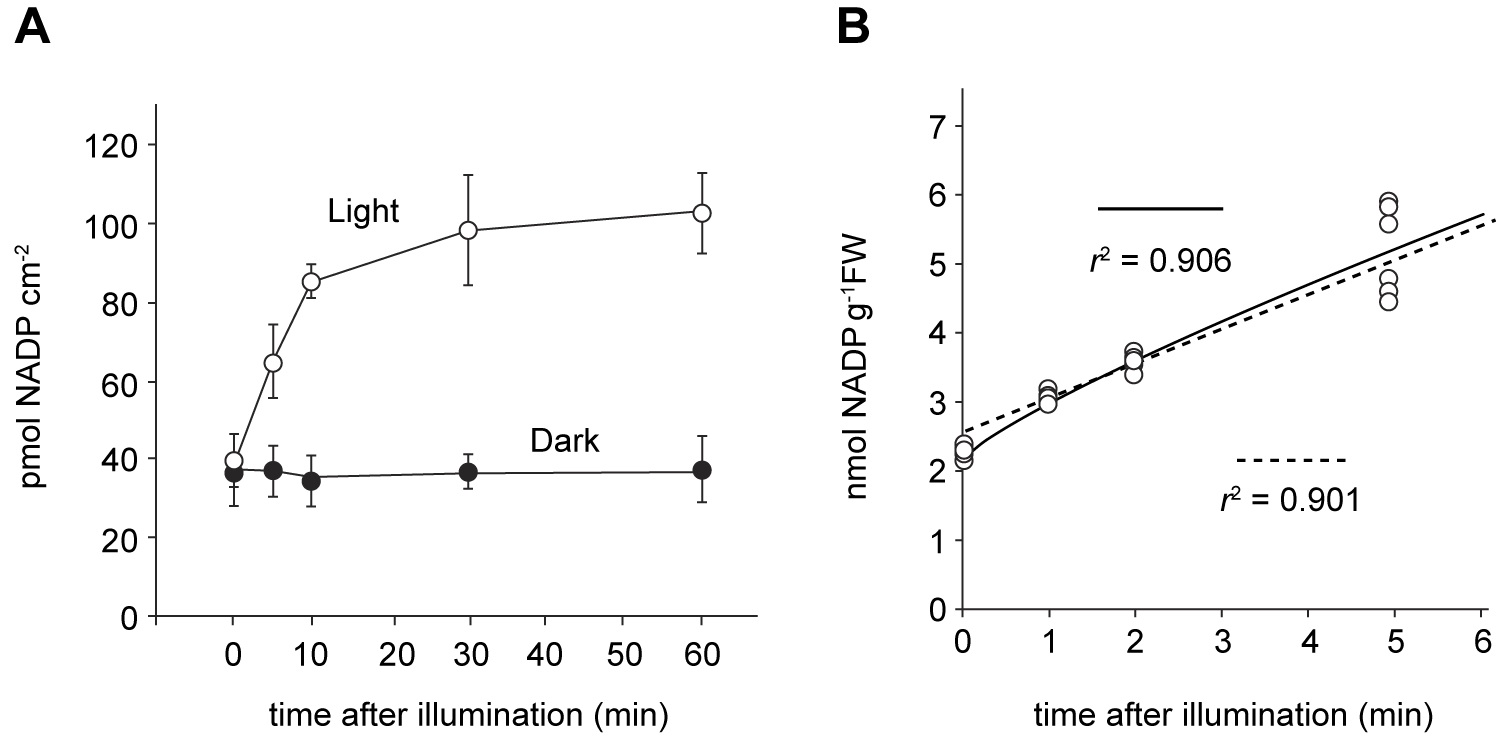
Figure 2. Light-driven NADP increase. A. Increase in the NADP content only occurs under illumination. B. Early NADP increase (for at least 5 min) was proportional to time. Result of straight fitting (dashed line) was quite comparable to the curve fitting (solid line) in the first 5 min of the illumination. - Curve fitting revealed some plausible chloroplastic NAD kinase activities at various time points (Figure 3, solid line). As we can see the initial activity was estimated to be over 50 nmol/h/g FW, but this activity declined rapidly in the first couple of minutes, and then slowly after about 5 min. On the other hand, simple fitting by straight line shows a representative estimation of the chloroplastic NAD kinase activity (Figure 3, dashed line). We expressed NAD kinase activities both as nmol/h/g FW and nmol NADP/h/mg protein, based on the protein amounts in the leaf discs. In previous reports, NADK2 activity has been estimated to be around 20 to 60 nmol/h/g FW (5 to 15 nmol/h/mg protein) (Takahashi et al., 2006 and 2009), which is consistent with the estimation from our newly developed assay.
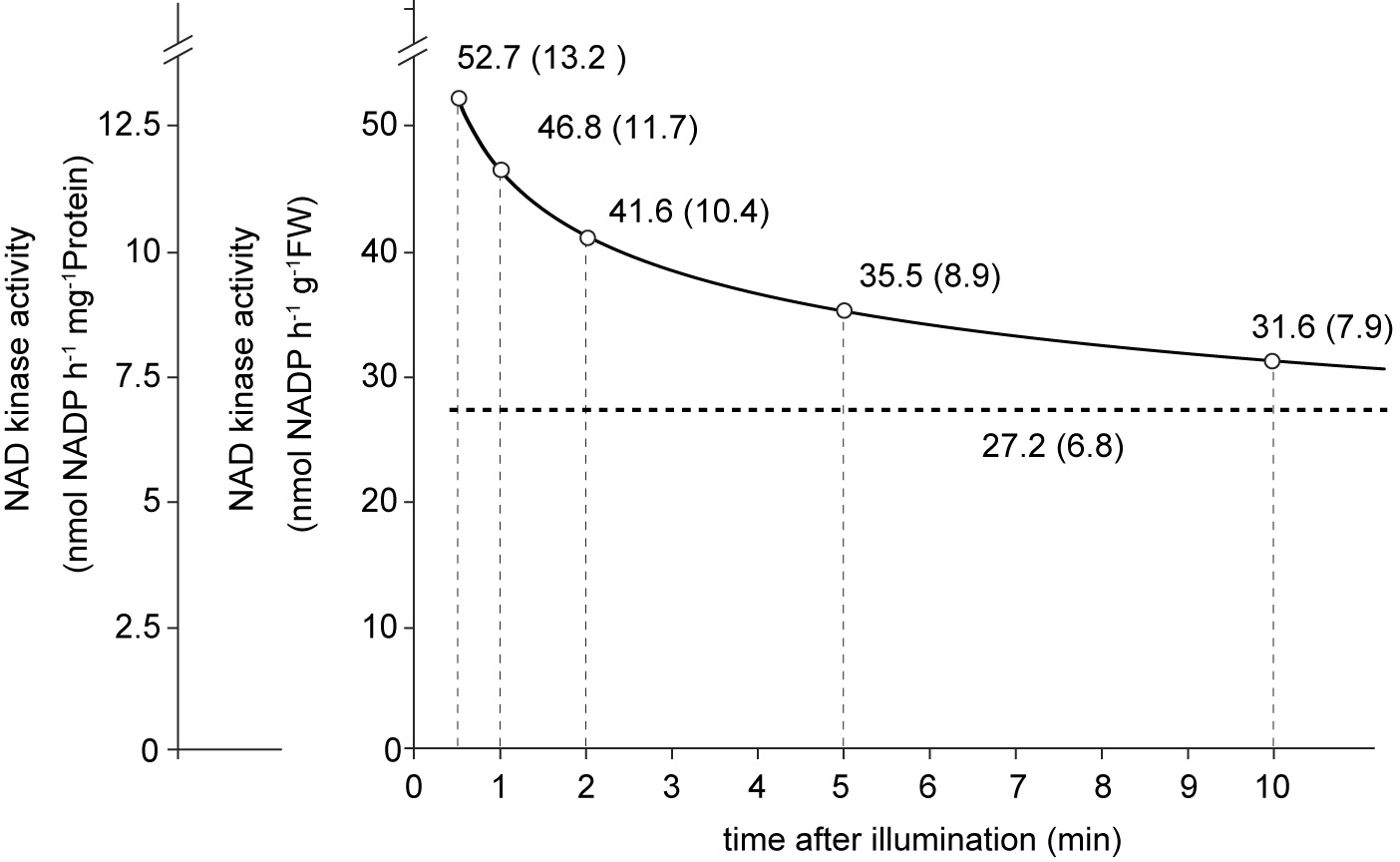
Figure 3. Estimation of chloroplastic NAD kinase activity. According to the quantification data, NAD kinase activity was estimated in nmol/h/g FW, and nmol/h/mg Protein. Values on the solid line were calculated from curve fitting, while the value under the dashed line was calculated from straight fitting. - Besides the NAD kinase activity, NADP synthesis is also determined by other factors (for example the NAD substrate and ATP) and therefore we believe that not only the in vitro, but also the newly developed assay will help to deduce which factors are conditionally involved in the regulation of NADP synthesis under various environmental signals.
Recipes
- Protein extraction buffer
50 mM HEPES/KOH, pH 8.0
10 mM MgCl2
1 mM CaCl2
1 mM PMSF
One tablet of cOmpleteTM, Mini Protease Inhibitor Cocktail per 10 ml - Reaction buffer
0.5 M HEPES/KOH, pH 8.0
100 mM MgCl2 - Luciferin detection reagent
Prepared fresh in the morning of the experiment, and stored at approximately 23 to 25 °C
Reconstruction Buffer, included in NADP/NADPH-GloTM Assay Kit
Lyophilized Luciferin, included in NADP/NADPH-GloTM Assay Kit - NADP/NADPH Glo detection reagent
Luciferin detection reagent, as described above
Reductase, included in NADP/NADPH-GloTM Assay Kit
Reductase Substrate, included in NADP/NADPH-GloTM Assay Kit
NADP Cycling Enzyme, included in NADP/NADPH-GloTM Assay Kit
NADP Cycling Substrate, included in NADP/NADPH-GloTM Assay Kit
Acknowledgments
This protocol was adapted and modified from the previous works by Takahashi et al. (2009) and Hashida et al. (2018). This work was supported by the JSPS KAKENHI Grant Number-17H05714 and 19H04715 to M.K.Y.
Competing interests
The authors declare that here are no conflicts of interest.
References
- Berrin, J. G., Pierrugues, O., Brutesco, C., Alonso, B., Montillet, J. L., Roby, D. and Kazmaier, M. (2005). Stress induces the expression of AtNADK-1, a gene encoding a NAD(H) kinase in Arabidopsis thaliana. Mol Genet Genomics 273(1): 10-19.
- Chai, M. F., Chen, Q. J., An, R., Chen, Y. M., Chen, J. and Wang, X. C. (2005). NADK2, an Arabidopsis chloroplastic NAD kinase, plays a vital role in both chlorophyll synthesis and chloroplast protection. Plant Mol Biol 59(4): 553-564.
- Dell’ Aglio, E., Giustini, C., Kraut, A., Couté, Y., Mazars, C., Matringe, M., Finazzi, G. and Curien, G. (2019). Identification of the Calmodulin-dependent NAD+ kinase sustaining the elicitor-induced oxidative burst in plants. bioRxiv: 521658.
- Delumeau, O., Renard, M. and Montrichard, F. (2000). Characterization and possible redox regulation of the purified calmodulin-dependent NAD+ kinase from Lycopersicon pimpinellifolium. Plant Cell Environ 23(11): 1267-1273.
- Hashida, S. N., Itami, T., Takahara, K., Hirabayashi, T., Uchimiya, H. and Kawai-Yamada, M. (2016). Increased rate of NAD metabolism shortens plant longevity by accelerating developmental senescence in Arabidopsis. Plant Cell Physiol 57(11): 2427-2439.
- Hashida, S. N. and Kawai-Yamada, M. (2019). Inter-organelle nad metabolism underpinning light responsive NADP dynamics in plants. Front Plant Sci 10: 960.
- Hashida, S. N., Miyagi, A., Nishiyama, M., Yoshida, K., Hisabori, T. and Kawai-Yamada, M. (2018). Ferredoxin/thioredoxin system plays an important role in the chloroplastic NADP status of Arabidopsis. Plant J 95(6): 947-960.
- Tai, L., Li, B. B., Nie, X. M., Zhang, P. P., Hu, C. H., Zhang, L., Liu, W. T., Li, W. Q. and Chen, K. M. (2019). Calmodulin is the fundamental regulator of NADK-mediated NAD signaling in plants. Front Plant Sci 10: 681.
- Takahashi, H., Takahara, K., Hashida, S. N., Hirabayashi, T., Fujimori, T., Kawai-Yamada, M., Yamaya, T., Yanagisawa, S. and Uchimiya, H. (2009). Pleiotropic modulation of carbon and nitrogen metabolism in Arabidopsis plants overexpressing the NAD kinase2 Gene. Plant Physiol 151(1): 100-113.
- Takahashi, H., Watanabe, A., Tanaka, A., Hashida, S. N., Kawai-Yamada, M., Sonoike, K. and Uchimiya, H. (2006). Chloroplast NAD kinase is essential for energy transduction through the Xanthophyll cycle in photosynthesis. Plant Cell Physiol 47(12): 1678-1682.
- Tezuka, T. and Yamamoto, Y. (1975). Photoactivation of NAD kinase through phytochrome: Phosphate donors and cofactors. Plant Physiol 56(6): 728-730.
- Turner, W. L., Waller, J. C., Vanderbeld, B. and Snedden, W. A. (2004). Cloning and characterization of two NAD kinases from Arabidopsis. identification of a calmodulin binding isoform. Plant Physiol 135(3): 1243-1255.
Article Information
Copyright
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Ishikawa, Y., Kawai-Yamada, M. and Hashida, S. (2020). Measurement of Chloroplastic NAD Kinase Activity and Whole Tissue NAD Kinase Assay. Bio-protocol 10(1): e3480. DOI: 10.21769/BioProtoc.3480.
Category
Plant Science > Plant biochemistry > Metabolite
Plant Science > Plant physiology > Metabolism
Biochemistry > Other compound > NAD+/NADH
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









