- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Semi-automated Model to Accurately Counting Sympathetic Nervous Fibers
Published: Vol 9, Iss 24, Dec 20, 2019 DOI: 10.21769/BioProtoc.3454 Views: 4678
Reviewed by: Ehsan KheradpezhouhGeoffrey C. Y. LauAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

The Mouse Social Frailty Index (mSFI): A Standardized Protocol
Charles W. Collinge [...] Alessandro Bartolomucci
Apr 20, 2025 1809 Views

A One-Step Mouse Model of Parkinson’s Disease Combining rAAV-α-Synuclein and Preformed Fibrils of α-Synuclein
Santhosh Kumar Subramanya [...] Poonam Thakur
Dec 5, 2025 1644 Views

A Low-Stress, Long-Duration Stable Tail Vein Catheterization and Precise Drug Delivery Protocol for Awake, Freely Moving Mice
Yunshuang Ye [...] Jun Fang
Feb 5, 2026 80 Views
Abstract
In recent years, the role of sympathetic nervous fibers in chronic inflammation has become increasingly evident. At the onset of inflammation, sympathetic activity is increased in the affected tissue. However, sympathetic fibers are largely absent from chronically inflamed tissues. Apparently, there is a very dynamic relationship between sympathetic innervation and the immune system in areas of inflammation, and hence a rapid and easy method for quantification of nerve fiber density of target organs is of great value to answer potential research questions. Sympathetic nerve ends lie in close proximity to immune cells in lymphoid tissues and lymphoid cells are equipped with catecholamine receptors. Catecholamines such as dopamine and adrenaline are secreted by sympathetic nervous fibers and can influence immune cell activity directly. Thereby the sympathetic nervous system immediately participates in the regulation of inflammation. Changes in innervation density could therefore indicate dysregulation of inflammatory processes. Currently, nervous fiber densities are either determined by tedious manual counting, which is not suitable for high throughput approaches, or by expensive automated processes relying on specialized software and high-end microscopy equipment. Usually, tyrosine hydroxylase (TH) is used as the marker for sympathetic fibers. In order to overcome the current quantification bottleneck with a cost-efficient alternative, an automated process was established and compared to the classic manual approach of counting TH-positive sympathetic fibers. Since TH is not exclusively expressed on sympathetic fibers, but also in a number of catecholamine-producing cells, a prerequisite for automated determination of fiber densities is to reliably distinguish between cells and fibers. Therefore, an additional stain using peripherin which is exclusively expressed in nervous fibers as a secondary marker was established. This new and simple method can be used as a high-throughput approach to reliably and quickly estimate sympathetic nervous system (SNS) nerve fiber density in target tissues.
Keywords: Peripheral nervous systemBackground
In order to provide less time-consuming alternatives to the tedious process of manually counting nervous fibers in tissues of interest and to stream line quantification and characterization of nervous fibers, automated and semi-automated processes have been developed and deployed as early as 1979. These processes require special equipment, such as array processors or specialized graphics ports and software, which is highly cost-intensive and often adapted to only one particular purpose (Frykman et al., 1979; Auer, 1994). To allow a more cost-efficient analysis of overall innervation in several target tissues, a semi-automated counting method was established by us. It is based upon several macros programmed for ImageJ using a basic fluorescence microscopy set-up. The working principle of the macros is demonstrated in Figure 1.
Usually, tyrosine hydroxylase (TH) is used as a marker of sympathetic fibers. TH catalyzes the conversion from L-tyrosine to L-3,4-dihydroxyphenylalanine (L-DOPA), which represents the rate-limiting step of catecholamine synthesis (Nagatsu et al., 1964; Hufton et al., 1995). TH is ubiquitously expressed in sympathetic nervous fibers as well as in a multitude of other cells throughout most mammalian tissues. Due to the fact that TH is not exclusively expressed in nervous fibers, we decided to introduce a counterstain to improve the distinction between TH-positive sympathetic fibers and TH-positive cells. The neurofilament peripherin is ubiquitously and exclusively expressed in nervous fibers and is therefore an excellent candidate for double stainings (Portier et al., 1983). We hypothesized, that with peripherin and TH co-staining, TH-positive fibers will be distinguished from TH-positive cells. Fibers will be discernible by the co-localization of peripherin and TH, while all other TH signals originate from TH-positive cells. If high-end technology required for automated counting processes is not available, fiber density is usually determined by manually counting visible TH-positive fibers in 17 high power fields (HPF), according to published methodology (Straub et al., 2008). However, this is a time-consuming and observer-dependent process. We present in this work a simple, high-throughput, automated screening method of sympathetic fiber density in tissues.
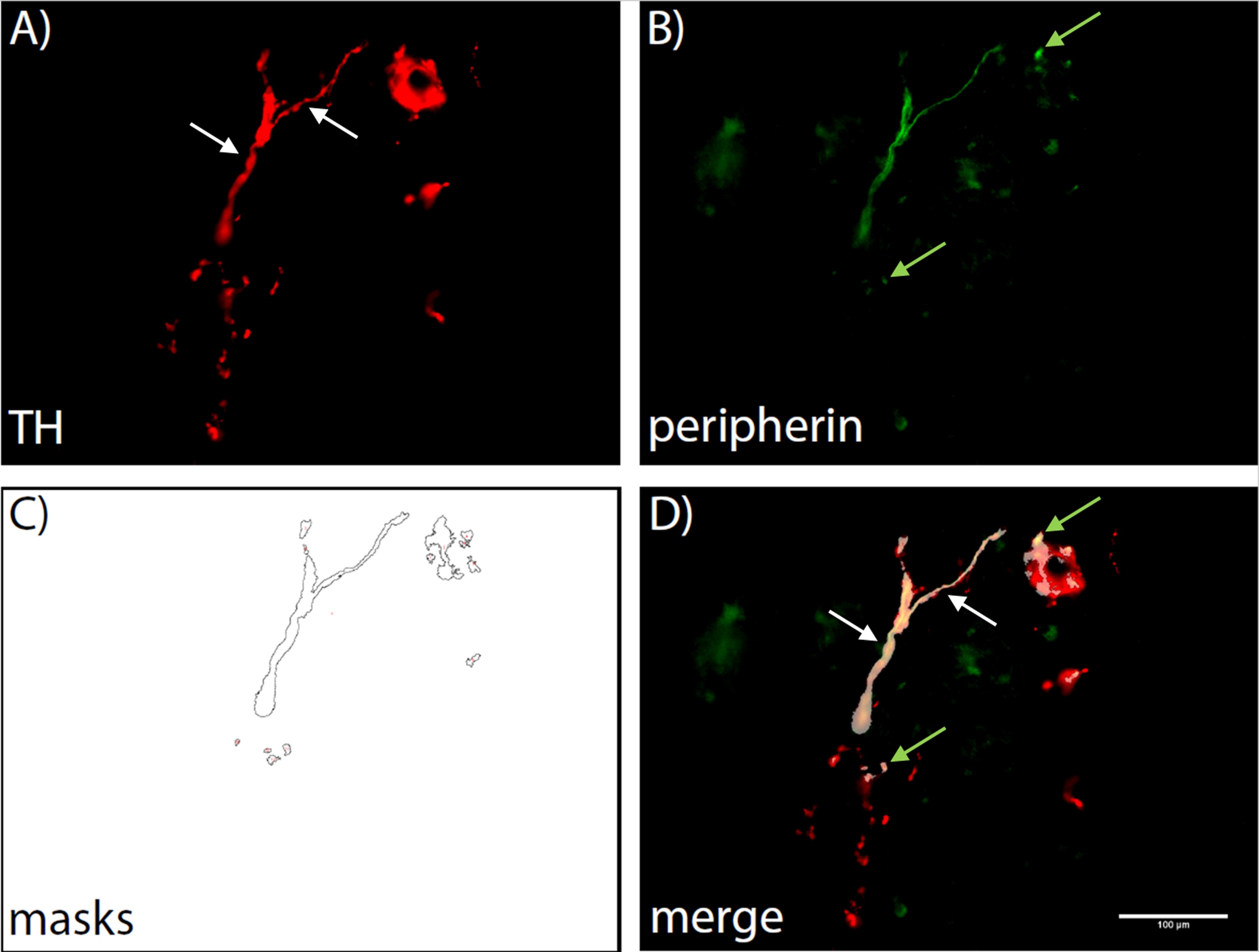
Figure 1. Demonstration of the automated counting process. A first macro creates a Region of interest (ROI) by automatically setting a threshold for TH positive areas (A). A second macro then lays the ROI over the corresponding peripherin stained image and creates masks wherever TH and peripherin staining are co-located (B, C). These masks are then counted and a list is displayed, showing the counts for each image that was analysed. A merge image is shown to illustrate the co-localization (D). White arrows indicate fibers that can be identified by eye, green arrows indicate those identified by co-localization only.
Materials and Reagents
- Pipette tips (Starlab, TipOne 1 ml, 200 µl, 10 µl, catalog numbers: S1111-6001, S1111-0000, S1111-3000)
- Microscopy cover glass (Engelbrecht, Cover Glass 24 mm x 60 mm, catalog number: K12460)
- Microtome blades (Feather, Disposable Microtome Blade, catalog number: C-35)
- Specimen molds (Sakura Finetek, Tissue-Tek Cryomold Intermediate 15 mm x 15 mm x 5 mm, catalog number: 4566)
- Microscopy slides (VWR, Adhesion slides SuperFrost Plus 75 mm x 25 mm, catalog number: 631-0108)
- PAP pen (Thermo Fisher, Super PAP Pen, catalog number: 008899)
- Erlenmayer flask (Brand, Erlenmayer flask 250 ml, catalog number: 92736)
- C57B/6J mice (Janvier Labs, C57B/6JRj, catalog number: C57B/6JRj)
- Goat Serum (NGS) (Agilent Dako, Goat Serum (Normal), Whole serum, 10 ml, catalog number: X090710-8), store at 4 °C
- Rabbit anti TH antibody (Merck, Anti-Tyrosine Hydroxylase Antibody, catalog number: AB152), store at -20 °C
- Chicken anti Peripherin antibody (Abcam, Anti-Peripherin antibody, catalog number: ab39374), store at -20 °C
- Goat anti Rabbit antibody (Invitrogen, Goat anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 594, catalog number: A-11037), store at 4 °C
- Goat anti Chicken antibody (Abcam, Goat Anti-Chicken IgY H&L (Alexa Fluor® 488), catalog number: ab150169), store at 4 °C
- Embedding Medium (Sakura Finetek, Tissue-Tek® O.C.T.TM Compound, 12 x 125 ml, catalog number: 4583), store at room temperature (RT)
- Triton X (Sigma-Aldrich, Triton TM X-100, catalog number: T8787), store at RT
- Paraformaldehyde (PFA) (Sigma-Aldrich, Paraformaldehyde, reagent grade, crystalline, catalog number: P6148), store at 4 °C
- Hydrochloric acid (Sigma-Aldrich, Hydrochloric acid 36.5-38.0%, BioReagent, for molecular biology, catalog number: H1758), store at RT
- Sodium Hydroxide (Merck, Sodium hydroxide 1.000 l, catalog number: 1099130001), store at RT
- Prolong Gold (Invitrogen, ProLongTM Gold Antifade Mountant with DAPI, catalog number: P36931), store at 4 °C
- Phosphate buffered Saline (Gibco, PBS (10x), pH 7.4, catalog number: 70011044), store at RT
- Liquid nitrogen (Linde, CRYOSPEED MED, catalog number: 4130919)
- Deionized H2O
- 3% PFA (250 ml) (see Recipes)
- PBS 1x (see Recipes)
- Blocking solution (see Recipes)
- Washing solution (see Recipes)
- Secondary antibody diluent solution (see Recipes)
Equipment
- Cryostat (Leica, CM3000 Research Cryostat)
- Microscope (Zeiss, Axioskop 2 plus)
- Camera (Nikon, DSV Vi1)
- PC (Intel Core i3-7100U CPU, 2.40 GHz, 8.00 GB RAM, Windows 10 Enterprise, 64-bit, Intel HD Graphics 620)
- Magnetic stir heater (IKA, IKAMAG, catalog number: RET)
- -80 °C freezer (Thermo Scientific, Forma 906, catalog number: 906)
- Electronic scale (Sartorius, electronic precision scale, catalog number: 1465 MP8)
- Shaker (Peqlab, Mini Rocker-Shaker, catalog number: MR-1)
- Micro pipettes (Eppendorf, Research plus variable 0,1-1 ml, 20-200 µl, 2-20 µl, 0,5-10 µl, catalog numbers: 3123000063, 3123000055, 3123000039, 3123000020)
Software
- ImageJ (NIH, https://imagej.nih.gov/ij/)
- NIS-Elements (Nikon, https://www.nikon.com/)
Procedure
- Tissue isolation and cryo fixation
- Isolate target tissue from mouse (e.g., spleen).
- Wash tissue once for 1-2 min in cold PBS 1x (4-8 °C).
- Put a basecoat of embedding medium in cryomold.
- Put the isolated organ/tissue into cryomold.
- Completely cover the tissue with embedding medium.
- Slowly lower the cryomold with the embedded tissue into liquid nitrogen.
- Wait until the whole block is frozen solid.
- Store embedded tissues at -80 °C until sectioning.
- Cryosectioning of the tissue
- Different tissues require different temperatures within the cryostat for optimal sections (depends on fat content, tissue density, etc.; a list of temperatures for different tissues is included in the manual for the Leica cryostat at: https://www.leicabiosystems.com/histology-equipment/cryostats/leica-cm3050-s/).
- Make sure the microtome blade is not dull (if marks or damage are visible or sections do not come off the blade easily or are disrupted during the sectioning process, replacing the blade might be necessary).
- Place the cryomold with the embedded tissue in the cool chamber of the cryostat for at least 30 min to allow equilibration of temperatures.
- When the embedded tissue has reached “sectioning temperature”, glue it to the specimen holder (part of the cryostat) using embedding medium. Make sure the tissue block is fixed and evenly glued to the specimen holder.
Note: Embedding medium might solidify rapidly on the cold specimen holder. - Wait for about 10 min to allow the embedding medium to completely solidify.
- Set the Cryostat to 100 µm steps and start trimming at 100 µm after 3 sections reduce trimming thickness to 50 µm. Perform each trimming step for 3 sections until you reach the desired thickness of the sections (in this case 3-5 µm because the thinner the sections are, the clearer the images will be later. Thicker sections cause images to be more diffuse).
Note: If there is only very little tissue to begin with, start trimming at thinner sections, or try sectioning the desired thickness right away. - Once reproducible sections of the desired thickness are produced, start collecting them by adhering them to microscopy slides.
- Leave the microscopy slides with the adherent tissue sections at room temperature (RT) to dry for at least 5 min.
- Fixation of sections
- Draw a circle of hydrophobic paraffin around the sections using PAP pen.
- Let the circles dry for about 1-2 min.
- Add about 100-300 µl of 3% PFA (Recipe 1) to each circle (liquid should not flow over the edges of the circles).
- Fixate tissue sections for 15 min at RT.
- Shake the PFA off of the slides and wash slides in a cuvette with PBS 1x for 10 min.
- Immunofluorescence staining
- Block (to reduce unspecific binding of antibodies) by adding blocking solution (Recipe 3) to each circle and incubate for 1 h at 4 °C.
- Aspirate the slides and add the primary antibodies (Rabbit anti TH antibody and Chicken anti Peripherin antibody) diluted 1:200 in blocking solution. Incubate overnight at 4 °C.
- Wash with washing solution (Recipe 4) for 10 min on a shaker at very low RPM (20-30 RPM). After 10 min, place the slides in fresh Washing buffer. Repeat wash for a total of three times.
Note: Work in the dark from this point to avoid bleaching of fluorophores. - Add secondary antibodies (Goat anti Rabbit antibody labeled with Alexa Fluor 594 and Goat anti Chicken antibody labeled with Alexa Fluor 488) diluted 1:1000 in secondary antibody diluent solution (Recipe 5) and incubate for 2 h at RT.
- Wash with PBS 1x for 10 min on a shaker at very low RPM (20-30 RPM). After 10 min, place the sections in fresh washing buffer. Repeat wash for a total of three times.
- Add 100-300 µl of Prolong Gold to each circle and cover the slide with a cover glass. Wipe off excess mounting medium with a paper tissue.
- Leave the slides at RT overnight to allow the Prolong Gold to cure.
- After the mounting medium has cured, store the slides at 4 °C until image acquisition.
- Image acquisition
- Capture individual images for TH, Peripherin and DAPI with 400 fold magnification (In this case 40x Neofluotar Zeiss Objective, the macros have been written for images of 400 fold magnification. If different magnifications are used, the particle size in the second macro needs to be adjusted. The macros have not been tested with images from more advanced microscopy setups, such as confocal systems. The higher signal to noise ratio could possibly lead to more precise fiber counts since thresholding is expected to be more precise).
- Create separate folders for TH, Peripherin and DAPI.
- Number the corresponding images accordingly when saving (e.g., TH1, Peripherin1, DAPI1; TH2, Peripherin2, DAPI2…).
- Automated fiber counting
- To add the counting macros to ImageJ (see Figure 2):
- Open ImageJ and click Plugins -> New -> Macro.
- Copy each macro from the supplement (Macros can be found in Supplement File and add the text to the empty new macro window.
- Save each macro where you will find it again and name them either TH.txt or Peripherin.txt.
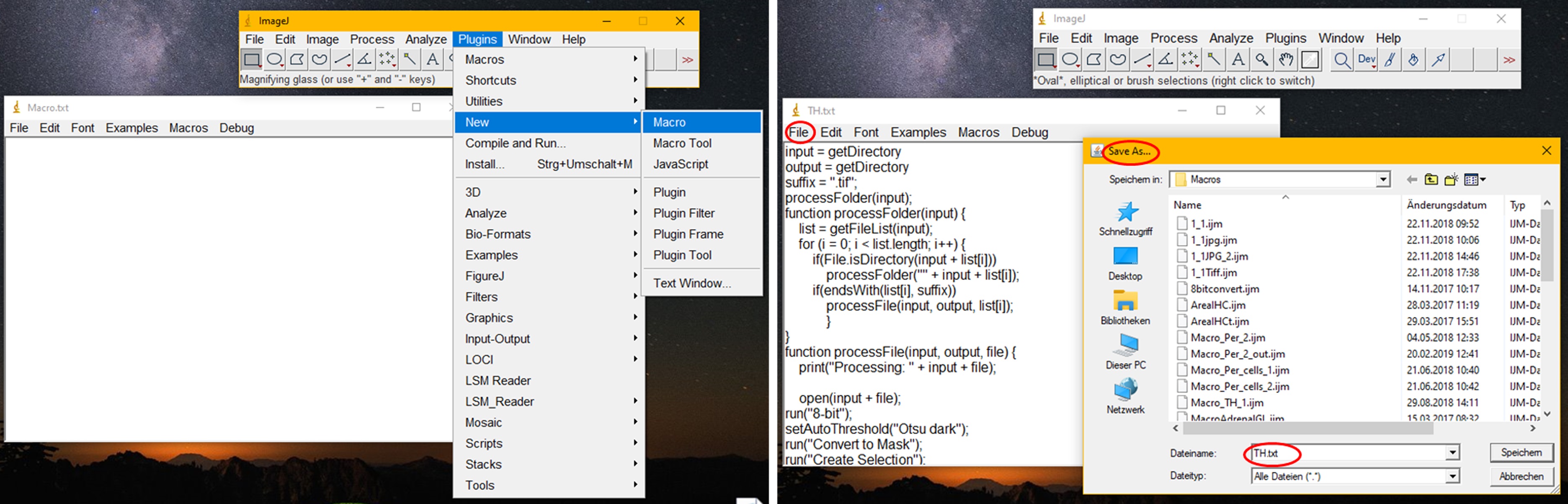
Figure 2. Instructions to add new macros to ImageJ. In order to install a new macro from text, open ImageJ, click on “Plugins” and click on “New” in the drop down menu. Then choose macro. A new window will open. You can now copy the code from the supplement into the new window. After the text was inserted, click “File”, “save as” and save the macros under a suitable name (in this case TH.txt and Peripherin.txt). Do not forget to add “.txt” to the file names, otherwise ImageJ will not be able to load the macros. - To run the macros, click Plugins -> Macros -> Run. Select the TH macro and run it for the TH folder to create regions of interest (ROIs) for TH positive areas (see Figure 3).
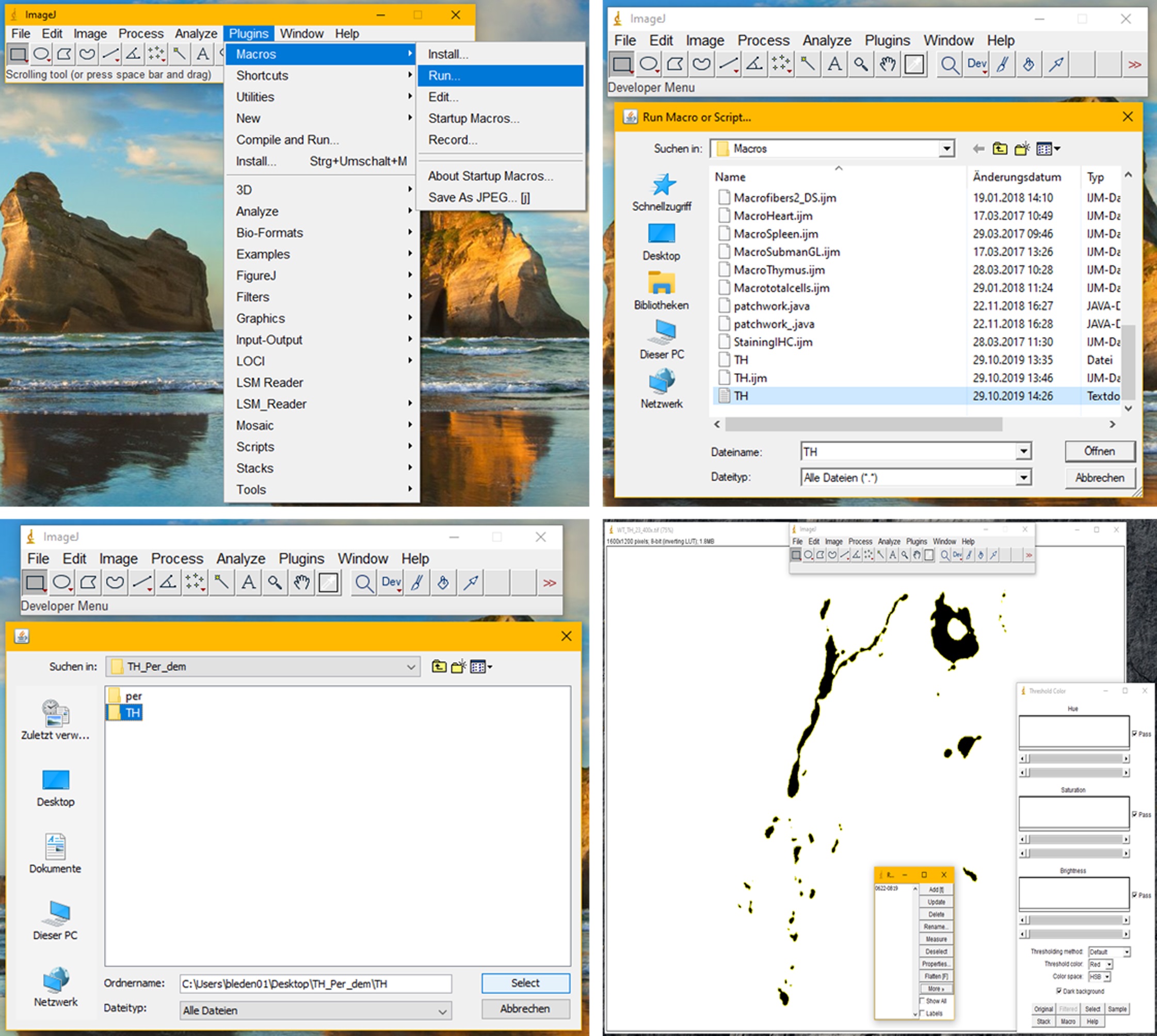
Figure 3. Instructions to run the first macro. The first macro is used to create ROIs of TH positive areas in the images in one folder. To run the macro, click “Plugins”, “Macros” and “Run”. Load the macro from where you saved it in step F1c. The Macro will ask you for an input and an output folder. Select the folder that contains the images of your TH stainings for both. The macro will then create an ROI for each of the images in the selected folder. The macro automatically sets a threshold for each image using the “Otsu dark” function of the ImageJ “Auto Threshold”. ROIs are then created based on the thresholds. In the ROI manager click on “more” and “save” to save the list to a location where you can easily retrieve it. If you are going to continue immediately with the second macro, you can leave the ROI manager open. Close the “Log” window, the “Threshold Color” window and all the images, that were opened by the macro (each of the images from the selected folder will be opened on the desktop). - Save the ROI list.
- Click Plugins -> Macros -> Select and run the Peripherin macro for the folder containing the peripherin images. This will scan for Peripherin positive areas within the TH positive ROIs (see Figure 4).
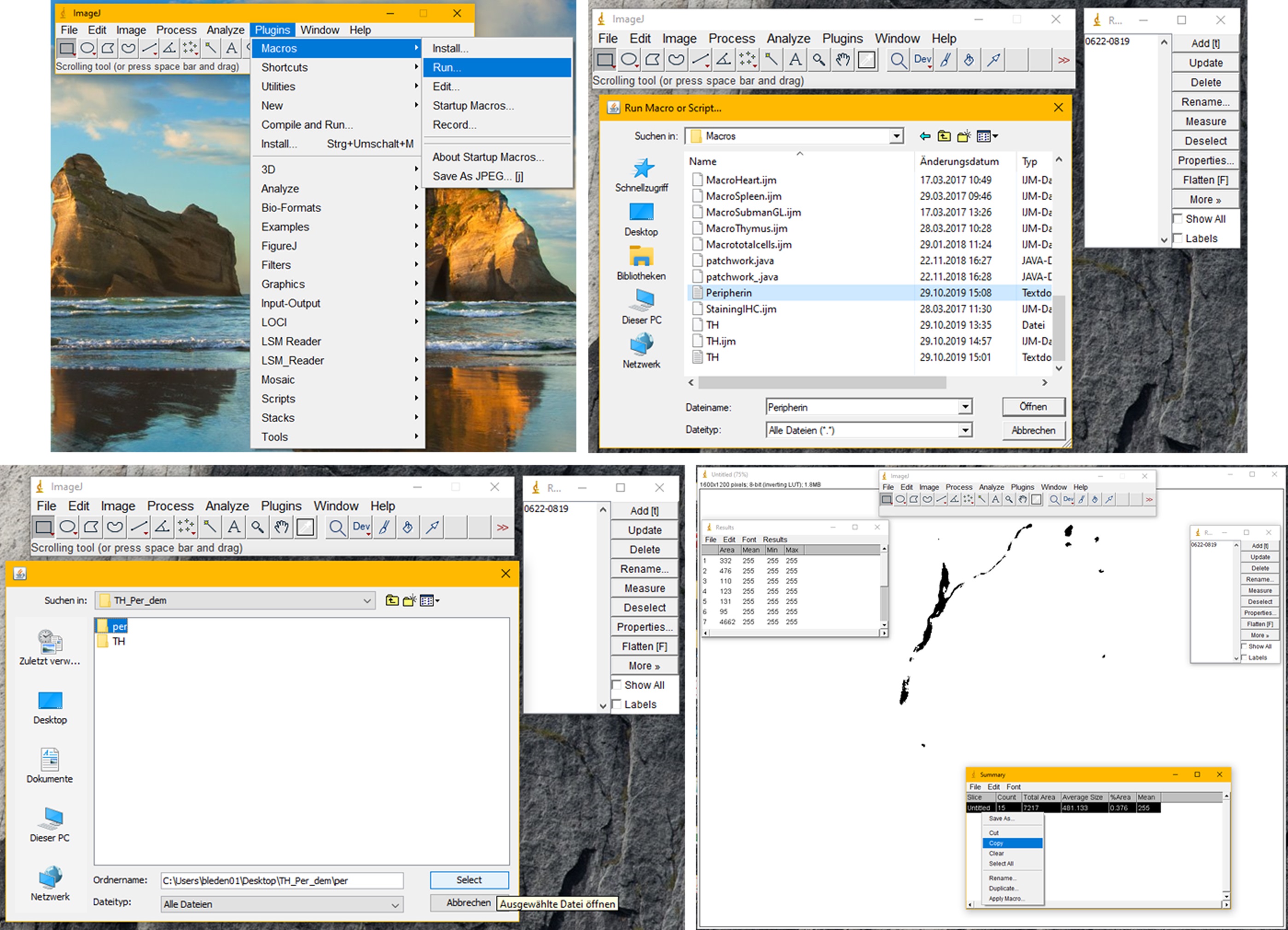
Figure 4. Instructions to run the second macro. The second macro is used to count peripherin positive particles within the TH positive ROIs. Before you run the second macro, make sure the ROI manager with the corresponding ROIs is open. Then run the macro by clicking “Plugins”, “Macros”, “Run” and selecting the Peripherin macro. The macro will once again ask you for an input and an output folder. This time choose the folder containing the images of your peripherin stainings for both. The macro automatically sets a threshold for each image using the “Default dark” function of the ImageJ “Auto Threshold”. Masks are created based on the threshold and these masks are counted by the “Analyse Particles” tool of ImageJ. Only masks of a size above 23 pixels are counted. If a lower magnification is used, the size needs to be decreased in the code. In the “Summary” window, the macro will create a list of all images within the selected folder showing the number of particles that were counted in each of them. All items on the list can be marked by pressing “ctrl-A” and copied into a data processing software by left-clicking and choosing copy. After the list has been copied, close ImageJ completely. - A count of particles for each image (image 1, image 2, image 3…) will be listed. Copy the list into the software you want to use for statistical analysis.
- Analyze data with software of your choice.
The method has been validated in comparison to manually counting the fibers in the original publication by Bleck et al. (2019). For examples of images from submandibular gland and heart sections with their corresponding counts see Figure 5.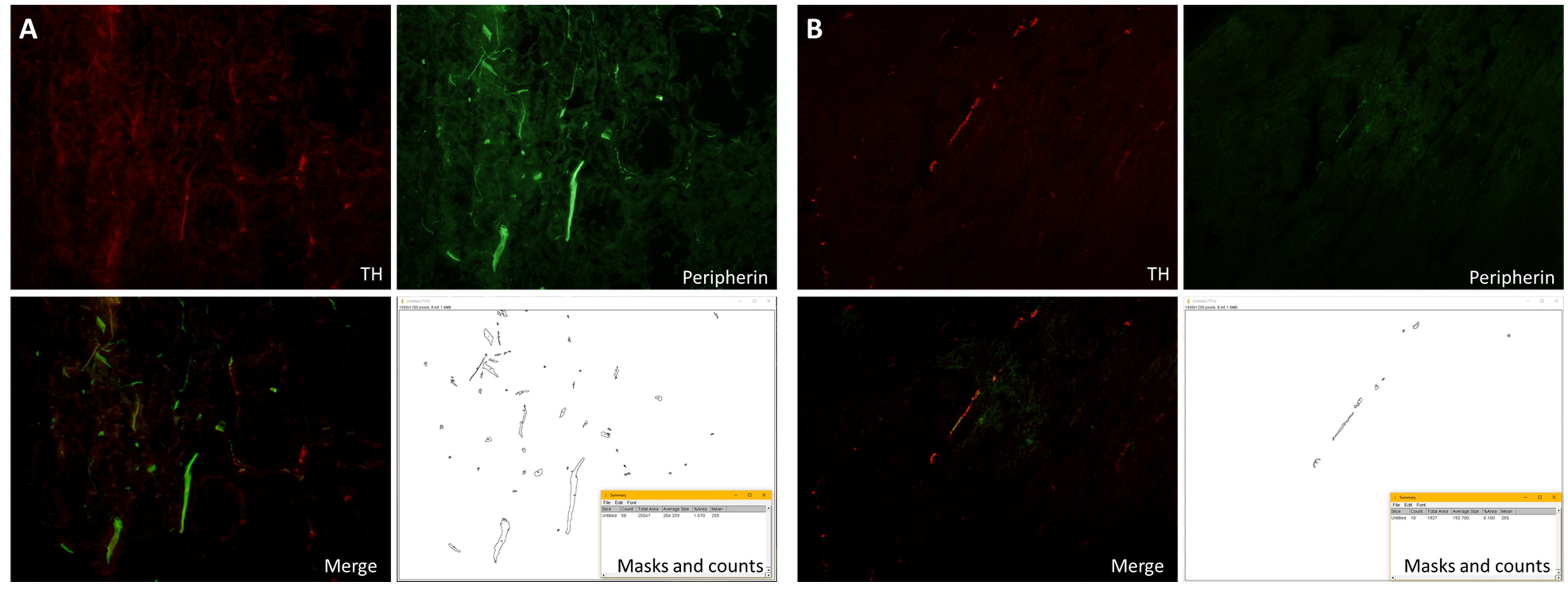
Figure 5. Examples of counts from images of submandibular gland and heart sections. A shows the staining and the counts from a submandibular gland section. The submandibular gland is densely innervated with SNS fibers. In comparison, B shows the staining and the result from a much less innervated heart section.
- To add the counting macros to ImageJ (see Figure 2):
Recipes
- 3% PFA (250 ml)
Pre-heat about 200 ml of de-ionized (DI) H2O to 60 °C in an Erlenmayer flask on a magnetic stir heater.- Weigh 7.5 g of of PFA in a 50 ml reaction tube
- Add about 40 ml of pre-heated H2O to the PFA and mix well
- Add the mixture to the rest of the H2O and keep stiring it
- Add 0.75 ml of 5 M NaOH
- Keep stiring, wait till the solution becomes clear
- Add 25 ml of 10x PBS
- Add 300 µl of concentrated HCl
- Adjust pH to 7 (adding either HCl or NaOH)
- Fill up to 250 ml with DI H2O
- Let the solution cool before tissue fixation
- Keep solution at 4 °C for about a week or prepare aliquots to freeze away at -20 °C for later use (Do not use after thawing more than once)
- PBS 1x
Add 1 volume of PBS 10x to 9 volumes of deionized H2O - Blocking solution
Add 2% NGS and 0.3% Triton X to the required volume of PBS 1x - Washing solution
Add 0.3% of Triton X to the required volume of PBS 1x - Secondary antibody diluent solution
Add 2% of NGS to the required volume of PBS 1x
Acknowledgments
This protocol was established for the original research paper “Introduction and validation of a new semi-automated method to determine sympathetic fiber density in target tissues” (Bleck et al., 2019).
Ethics
C57B/6J mice used to establish this protocol were bred and sacrificed for organ extraction at the animal experiment facility of the University of Düsseldorf (ZETT). All methods were carried out in accordance with the approved guidelines under the project ID O 57/15.
References
- Auer, R. N. (1994). Automated nerve fibre size and myelin sheath measurement using microcomputer-based digital image analysis: theory, method and results. J Neurosci Methods 51(2): 229-238.
- Bleck, D., Ma, L., Erdene-Bymbadoo, L., Brinks, R., Schneider, M., Tian, L. and Pongratz, G. (2019). Introduction and validation of a new semi-automated method to determine sympathetic fiber density in target tissues. PLoS One 14(5): e0217475.
- Frykman, G. K., Rutherford, H. G. and Neilsen, I. R. (1979). Automated nerve fiber counting using an array processor in a multi-minicomputer system. J Med Syst 3(1-2): 81-94.
- Hufton, S. E., Jennings, I. G. and Cotton, R. G. (1995). Structure and function of the aromatic amino acid hydroxylases. Biochem J 311 (Pt 2): 353-366.
- Nagatsu, T., Levitt, M. and Udenfriend, S. (1964). Tyrosine hydroxylase. The initial step in norepinephrine biosynthesis. J Biol Chem 239: 2910-2917.
- Portier, M. M., de Néchaud, B. and Gros, F. (1983). Peripherin, a new member of the intermediate filament protein family. Dev Neurosci 6(6): 335-344.
- Straub, R. H., Rauch, L., Fassold, A., Lowin, T. and Pongratz, G. (2008). Neuronally released sympathetic neurotransmitters stimulate splenic interferon-gamma secretion from T cells in early type II collagen-induced arthritis. Arthritis Rheum 58(11): 3450-3460.
Article Information
Copyright
© 2019 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Bleck, D., Erdene-Byambadoo, L., Brinks, R., Schneider, M. and Pongratz, G. (2019). Semi-automated Model to Accurately Counting Sympathetic Nervous Fibers. Bio-protocol 9(24): e3454. DOI: 10.21769/BioProtoc.3454.
Category
Neuroscience > Behavioral neuroscience > Animal model
Neuroscience > Nervous system disorders > Animal model
Neuroscience > Sensory and motor systems > Animal model
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








