- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Proximity Ligation Assay for the Investigation of the Intramolecular Interaction of ELMO1
(*contributed equally to this work) Published: Vol 9, Iss 23, Dec 5, 2019 DOI: 10.21769/BioProtoc.3449 Views: 4779
Reviewed by: Edgar Soria-GomezTongxing SongAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Determination of Dissociation Constants for the Interaction of Myosin-5a with its Cargo Protein Using Microscale Thermophoresis (MST)
Rui Zhou [...] Xiang-Dong Li
Feb 5, 2025 1738 Views

Cell-Sonar, an Easy and Low-cost Method to Track a Target Protein by Expression Changes of Specific Protein Markers
Sabrina Brockmöller [...] Simone Rothmiller
Feb 5, 2025 1657 Views

SiMPull-POP: Quantification of Membrane Protein Assembly via Single Molecule Photobleaching
Ryan J. Schuck [...] Rajan Lamichhane
Jan 5, 2026 287 Views
Abstract
Intramolecular interaction is a common mechanism that regulates protein activities. Conventionally, such interactions are investigated by classical in vitro biochemical assays. Here, we describe a protocol for studying the intramolecular interaction of cell motility and engulfment 1 (ELMO1) in mammalian cells by using proximity ligation assay (PLA). PLA is a specific and sensitive method that allows the observation of interacting proteins by target-specific antibody detection coupled to rolling circle amplification. ELMO1 is the regulatory subunit of ELMO1-dedicator of cytokinesis 180 (DOCK180) bipartite Rac1 guanine nucleotide exchange factor (GEF) which adopts a closed autoinhibitory conformation via an intramolecular interaction of its N-terminal ELMO inhibitory domain (EID) and C-terminal ELMO autoregulatory domain (EAD). In the assay, PLA signals are detected in cells transfected with ELMO11-315 and ELMO1315-727 fragments. Moreover, overexpression of FE65, a neuronal adaptor which has been shown to disrupt ELMO1 intramolecular interaction, reduces the PLA signals of the two ELMO1 fragments significantly. Together, our results demonstrate that PLA can be employed for studying protein intramolecular interaction.
Keywords: AutoinhibitionBackground
The activities of proteins in cells can be regulated by various mechanisms including intramolecular interaction (Miernyk and Thelen, 2008). Several protein-protein interaction assays have been adopted for studying intramolecular interaction such as co-immunoprecipitation and protein complementation assays (PCAs). Despite being widely employed, there are known limitations of these conventional assays. For instance, co-immunoprecipitation is not ideal for detecting low-affinity interaction. Even though PCAs provide good sensitivity, the reporter fragments may hinder the interaction of interest (Ohad et al., 2007; Miernyk and Thelen, 2008). Proximity ligation assay (PLA) is an innovative technology that allows in situ detection of protein-protein interactions. The protein targets are first labeled by their specific primary antibodies from different species, followed by the recognition of species-specific secondary antibodies conjugated to oligonucleotides (PLA probes). Proximity ligation of the oligonucleotides generates the templates for single-stranded rolling circle amplification. The resultant products are detected with fluorescence-labeled complementary oligonucleotide detection probes. PLA offers exceptional specificity and sensitivity as target-specific antibodies and single-stranded rolling circle amplification are involved. The resultant products are detected with fluorescence-labeled complementary oligonucleotide detection probes, and PLA signals can be visualized and quantified by using fluorescence microscopy (Weibrecht et al., 2010). Although the technology has been available for over a decade, the possibility of using PLA for investigating protein intramolecular interaction has not been explored.
Engulfment and cell motility 1 (ELMO1) is the regulatory subunit of the bipartite ELMO1-dedicator of cytokinesis 180 (DOCK180) guanine nucleotide exchange factor (GEF) for the activation of the small GTPase Rac1. ELMO1 possesses several distinct domains: a Ras binding domain (RBD), ELMO inhibitory domain (EID), ELMO domain, pleckstrin homology (PH) domain, ELMO autoregulatory domain (EAD) and a PXXP motif (Figure 1). Similar to many other GEFs, the activity of ELMO1 is modulated by the intramolecular interaction between its EID and EAD which allows ELMO1 to adopt a closed autoinhibitory conformation at the basal state (Patel et al., 2010) (Figure 1). The relief of ELMO1 autoinhibition is required for the targeting of the complex of ELMO1-DOCK180 to the plasma membrane where it stimulates Rac1 (Patel et al., 2011). However, the mechanism that alleviates the autoinhibition of ELMO1 remains elusive. We have recently reported that the neuronal adaptor protein FE65 interacts with ELMO1 EAD and to disrupt the interaction of ELMO11-315 (comprises of ELMO1 amino acid residues 1-315) and ELMO1315-727 (comprises of ELMO1 amino acid residues 315-727) fragments, which possess the EID and EAD respectively, by using both classical protein binding assay and PLA (Li et al., 2018). Therefore, we demonstrate for the first time that PLA can be applied for examining protein intramolecular interaction.
Here, we describe how to investigate the intramolecular interaction of ELMO1 EID and EAD by using PLA. The method may also be adopted for the investigation of other protein intramolecular interactions.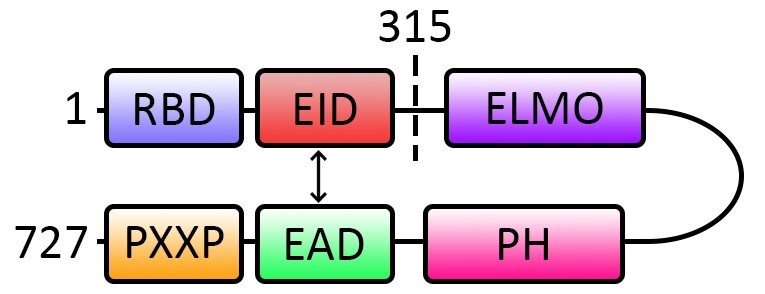
Figure 1. Schematic diagram shows the subdomains and the intramolecular interaction of ELMO1. At basal stage, the EAD and EID interact intramolecularly (indicated with double headed arrow) to form an autoinhibitory conformation. The disruption of the intramolecular interaction alleviates the autoinhibition for the subsequence Rac1 activation. Numbers denote the amino acid residue number in ELMO1. RBD: Ras-binding domain; EID: ELMO inhibitory domain; ELMO: ELMO domain; PH: Pleckstrin homology domain; EAD: ELMO autoregulatory domain; PXXP: PXXP motif.
Materials and Reagents
- SterilinTM Standard 90 mm Petri Dish (Thermo Scientific, catalog number: 101VIRR), stored at room temperature
- 12-well cell culture plate (SPL, catalog number: 30012), stored at room temperature
- Cover glass 18-mm circles (Thermo-Menzel, catalog number: CB00180RA1), stored at room temperature
- Hemocytometer (HBG, catalog number: 9010-01), stored at room temperature
- Microscope slides (Thermo Scientific, catalog number: 6776214), stored at room temperature
- HEK293 cells (ATCC, catalog number: CRL-1573), stored in liquid nitrogen
- Goat anti-ELMO1 antibody (Santa Cruz Biotechnology, catalog number: sc-21651), stored at 4 °C
- Mouse anti-FLAG antibody (Sigma-Aldrich, catalog number: F1804), stored at -20 °C
- Rabbit anti-β-tubulin antibody (Abcam, catalog number: AB6046)
- Donkey anti-rabbit IgG (H + L) highly cross-adsorbed secondary antibody, Alexa Fluor 488 (Invitrogen, catalog number: A21206), stored at 4 °C
- Fetal bovine serum (HyClone, catalog number: SV30160.03), stored at -20 °C
- pCMV-Tag2B vector (Aligent, catalog number: 211172), stored at -20 °C
- T4 DNA Ligase (1 U/μl) (Thermo Scientific, catalog number: 15224025), stored at -20 °C
- BamHI (10 U/μl) (Thermo Scientific, catalog number: ER0055), stored at -20 °C
- XhoI (10 U/μl) (Thermo Scientific, catalog number: ER0691), stored at -20 °C
- TIANprep Mini Plasmid Kit (TIANGEN, catalog number: DP103), stored at room temperature
- QuikChange II Site-Directed Mutagenesis Kit (Agilent, catalog number: 200523), stored at -20 °C
- Phusion High-Fidelity PCR Kit (Thermo Scientific, catalog number: F553S), stored at -20 °C
- UltraPureTM Agarose (Invitrogen, catalog number: 16500500), stored at room temperature
- TAE Buffer (Tris-acetate-EDTA) (50x) (Thermo Scientific, catalog number: B49), stored at room temperature
- UltraPureTM Ethidium Bromide, 10 mg/ml (Invitrogen, catalog number: 15585011), stored at room temperature
- 1 kb Plus DNA Ladder (New England Biolabs, catalog number: N3200), stored at -20 °C
- Luria Broth Base, powder (Invitrogen, catalog number: 12795084), stored at room temperature
- Clear nail polish
- LB Agar, powder (Invitrogen, catalog number: 22700041), stored at room temperature
- Ampicillin sodium salt (Sigma-Aldrich, catalog number: A0166), stored at 4 °C
- Kanamycin sulfate (Sigma-Aldrich, catalog number: 60615), stored at 4 °C
- Poly-D-lysine hydrobromide (Sigma-Aldrich, catalog number: P7280), stored at -20 °C
- DMEM (HyClone, catalog number: SH30021.01), stored at 4 °C
- Trypsin-EDTA 0.05% (Gibco, catalog number: 25300062), stored at -20 °C
- Opti-MEM reduced serum medium (Gibco, catalog number: 31985088), stored at 4 °C
- X-tremeGENETM 9 DNA transfection reagent (Sigma-Aldrich, catalog number: 6365779001), stored at 4 °C
- PBS (Gibco, catalog number: 10010023), stored at room temperature
- Paraformaldehyde (Sigma-Aldrich, catalog number: P6148), stored at 4 °C
- Triton X-100 (Anatrace, catalog number: T1001500ML), stored at room temperature
- Duolink® In Situ Red Starter Kit Mouse/Goat (Sigma-Aldrich, catalog number: DUO92103), stored at -20 °C
- 1x TAE buffer (see Recipes)
- Poly-D-lysine solution (see Recipes)
- 4% paraformaldehyde (freshly prepared) (see Recipes)
- 1x Ligation buffer (see Recipes)
- 1x Amplification red (see Recipes)
- Wash buffer A (see Recipes)
- Wash buffer B (see Recipes)
Equipment
- Water bath (Labnet, model: W1106)
- Heat block (Labnet, model: D1301-230V)
- UV transilluminator (Accuris, model: E3000)
- T100TM Thermal Cycler (Bio-rad)
- HerathermTM Compact Microbiological Incubators (37 °C, Thermo Scientific)
- FormaTM Series II 3110 Water-Jacketed CO2 Incubators (37 °C, 5% CO2, Thermo Scientific)
- Fluorescence microscope (Nikon, model: Eclipse Ni-U) equipped with QI2 high-resolution microscope camera (Nikon)
Software
- NIS-Elements research basic (Nikon)
- Prism (https://www.graphpad.com/scientific-software/prism/)
Procedure
- Generation of mammalian expression construct of ELMO11-315
- Perform site-directed mutagenesis to introduce a stop codon after ELMO1 residue 315 in pcDNA3-N-Myc-Elmo1 by using QuikChange II Site-Directed Mutagenesis Kit (Figure 2). The mutagenesis primers used are listed in Table 1.

Figure 2. Schematic diagram of ELMO1 fragment constructs. Site-directed mutagenesis is performed to create a stop codon after ELMO1 residue 315 in pcDNA3-N-Myc-Elmo1 to generate ELMO11-315 fragment. cDNA of ELMO1315-727 is amplified from pcDNA3-N-Myc-Elmo1 by using PCR and the product is inserted to pCMV-Tag2B for the expression of N-terminal FLAG tagged protein. The asterisk indicates the mutation site. Numbers denote the amino acid residue number in ELMO1.
Table 1. Primers used for generating ELMO1 fragment constructs. Primers 1 and 2 are used for the site-directed mutagenesis to introduce a stop codon to the pcDNA3-N-Myc-Elmo1. Primers 3 and 4 are used for the amplification of ELMO1315-727 fragment by using pcDNA3-N-Myc-Elmo1 as a template.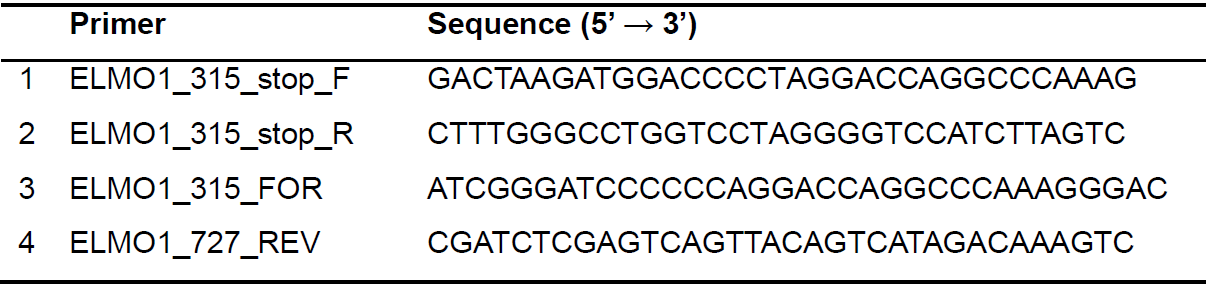
- Transform the mutagenesis product to DH5α competent E. coli cells by heat shock for 1 min at 42 °C.
- Plate the transformants on LB agar plates with 100 μg/ml ampicillin.
- Incubate the plates overnight in 37 °C incubator.
- Inoculate isolated colonies in 5 ml LB with 100 μg/ml ampicillin and grow overnight at 37 °C.
- Isolate the plasmid DNA by using TIANprep Mini Plasmid Kit.
- Perform DNA sequencing to confirm the introduction of the stop codon.
- Perform site-directed mutagenesis to introduce a stop codon after ELMO1 residue 315 in pcDNA3-N-Myc-Elmo1 by using QuikChange II Site-Directed Mutagenesis Kit (Figure 2). The mutagenesis primers used are listed in Table 1.
- Generation of mammalian expression construct of FLAG-ELMO1315-727
- Perform polymerase chain reaction (PCR) to obtain cDNA encoded for ELMO1315-727 fragment by using Phusion High-Fidelity PCR Kit with pcDNA3-N-Myc-Elmo1 as a template. Primers used are listed in Table 1.
- Resolve the PCR product by a 1% agarose gel with 0.2 μg/ml ethidium bromide in 1x TAE buffer.
- Visualize the PCR product by using a UV transilluminator and excise the PCR product (~1.2 kb) with a razor blade after electrophoresis.
- Purify the PCR product from gel by using MEGAquick-spinTM Plus Total Fragment DNA Purification Kit.
- Digest the purified ELMO1315-727 PCR product and pCMV-Tag2B, a vector with an N-terminal FLAG tag, with BamHI and XhoI for 2 h in a 37 °C water bath.
- Purify the digestion products by using MEGAquick-spinTM Plus Total Fragment DNA Purification Kit.
- Ligate the digested ELMO1315-727 fragment and linearized pCMV-Tag2B vector with T4 DNA ligase overnight in a 16 °C water bath.
- Transform the ligation product as stated in Steps A2-A6. 50 μg/ml Kanamycin should be used for selection.
- Perform DNA sequencing to confirm the successful insertion of the ELMO1315-727 encoding cDNA into pCMV-Tag2B.
- Cell preparation and transfection
- Coat 18-mm cover glasses with 5 μg/ml poly-D-lysine solution in 12-well cell culture plate overnight in a 37 °C incubator.
Note: Sterilize the cover glass before use. - Wash the cover glasses once with distilled water.
- Seed HEK293 cells on the pre-coated 18-mm cover glasses at 0.4 x 105 cells in 1 ml DMEM medium supplemented with 10% FBS.
- After 24 h, transfect the cells with mammalian expression constructs of myc-ELMO11-315 and FLAG-ELMO1315-727 together with either empty vector (EV), FE65 or FE65 K48A/R51A (FE65m, an ELMO1 binding defective FE65 mutant) using X-tremeGENETM 9 DNA transfection reagent in Opti-MEM reduced serum medium according to the manufacturer’s instruction (Table 2).
Table 2 Transfection combinations for studying the effect of FE65 in ELMO1 intramolecular interaction. HEK293 cells are co-transfected with either EV, FE65 or FE65m together with ELMO11-315 and ELMO1315-727 constructs in 1:1:1 ratio. All transfections receive the same amount of DNA. “+” denotes the presence of the plasmid while “-” denotes the absence of the plasmid.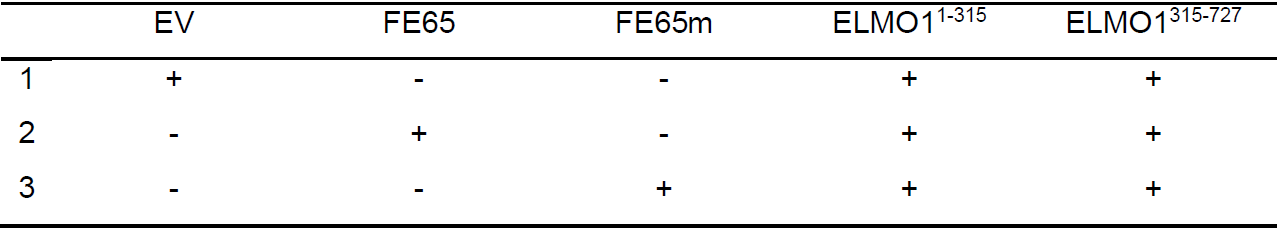
- Coat 18-mm cover glasses with 5 μg/ml poly-D-lysine solution in 12-well cell culture plate overnight in a 37 °C incubator.
- Cell fixing and proximity ligation assay
Note: Wash Buffer A, Wash Buffer B, PLA probe anti-goat MINUS, PLA probe anti-mouse PLUS, Antibody Diluent, Ligase, 5x Ligation Buffer, Polymerase, 5x Amplification Red and Duolink In Situ Mounting Media with DAPI are provided within Duolink® In Situ Red Starter Kit Mouse/Goat. Please see Figure 3 for the workflow of PLA.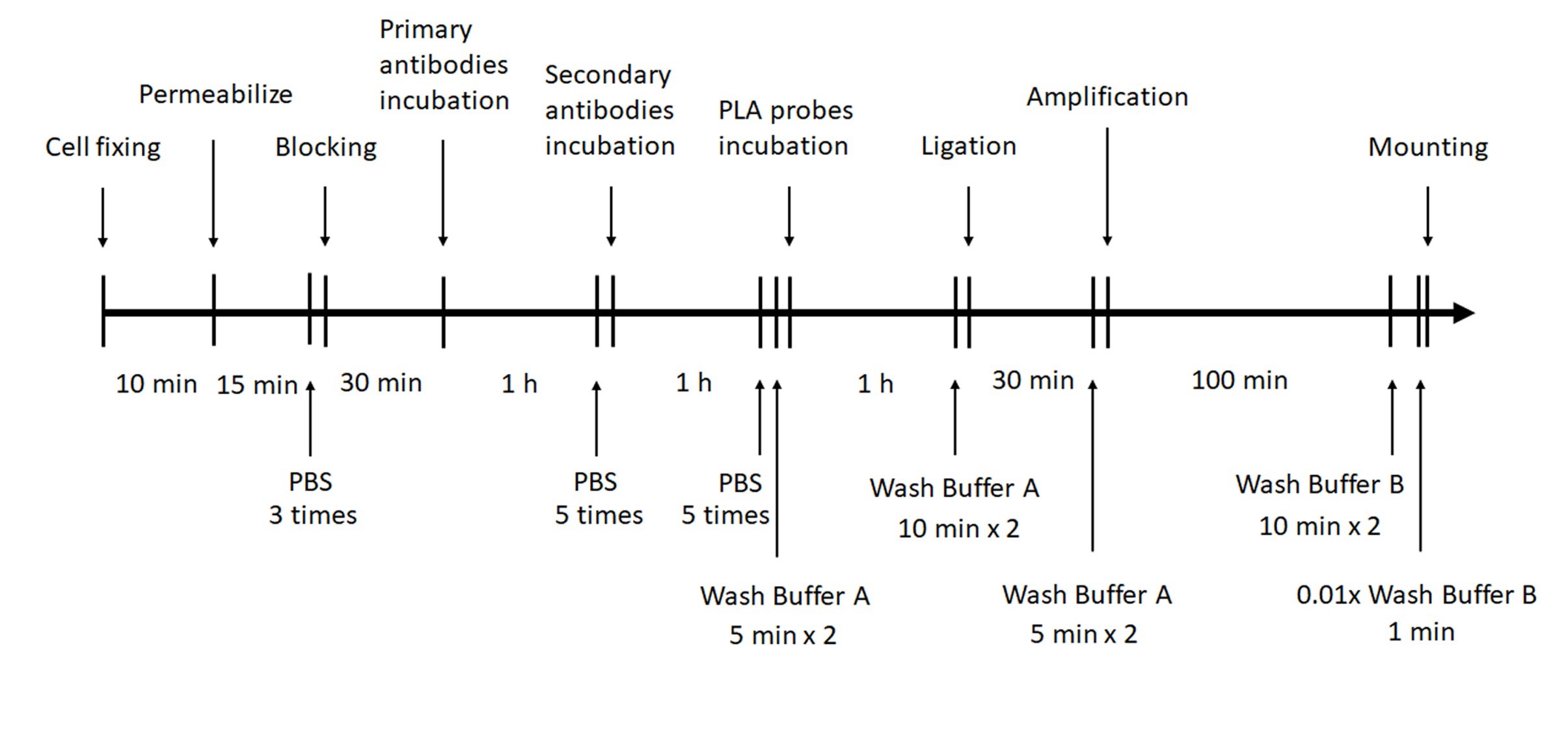
Figure 3. Timeline illustrates the workflow of PLA of ELMO1 fragments using Duolink® In Situ Red Starter Kit Mouse/Goat- After 24 h transfection, wash the cells once with PBS and fix the cells with 4% paraformaldehyde for 10 min at room temperature.
- Wash the fixed cells three times with PBS.
- Permeabilize the cells with 0.1% Triton X-100/PBS for 15 min at room temperature.
- Block the cells with 5% FBS/PBS for 30 min at room temperature.
- Dilute the goat anti-ELMO1 (recognizes myc-ELMO11-315) (1:100), mouse anti-FLAG (recognizes FLAG-ELMO1315-727) (1:1000) and rabbit anti-β-tubulin (1:200) antibodies in 5% FBS/PBS and stain the cells with 80 μl of diluted antibody mix for 1 h at room temperature.
Note: The concentrations of primary antibodies used are critical for obtaining high-quality PLA images and should be optimized. - Wash the cells five times with PBS.
- Dilute the Alexa Fluor 488 conjugated Donkey anti-rabbit IgG (1:1000) antibody in 5% FBS/PBS and stain the cells for 1 h in the dark at room temperature.
- Wash the cells five times with PBS.
- Wash the cells twice with Wash Buffer A for 5 min in the dark at room temperature.
- Add 80 μl PLA probe reaction mix to each cover glass and incubate in a humidified 37 °C incubator for 1 h.
Note: For each reaction, mix 16 μl of PLA probe anti-goat MINUS (Donkey anti-goat secondary antibody conjugated to oligonucleotide MINUS) with 16 μl of PLA probe anti-mouse PLUS (Donkey anti-mouse secondary antibody conjugated to oligonucleotide PLUS) in 48 μl of Antibody Diluent and incubate for 20 min at room temperature before use. - Wash the cover glasses twice with Wash Buffer A for 5 min in the dark at room temperature.
- Add 80 μl Ligation reaction mix to each cover glass and incubate in a humidified 37 °C incubator for 30 min.
Note: For each reaction, add 2 μl Ligase to 78 μl 1x Ligation buffer. - Wash the cover glasses twice with Wash Buffer A for 5 min in the dark at room temperature.
- Add 80 μl Amplification reaction mix to each cover glass and incubate in humidified 37 °C incubator for 100 min.
Note: For each reaction, add 1 μl Polymerase to 79 μl 1x Amplification Red. - Wash the cover glasses twice with Wash Buffer B for 10 min in the dark at room temperature.
- Wash the cover glasses once with 0.01x Wash Buffer B for 1 min in the dark at room temperature.
- Mount the cover glasses on microscope slides with a minimal amount of Duolink In Situ Mounting Media with DAPI.
- Seal the edge of the mounted cover glasses with clear nail polish.
- Capture the cell images with a 60x water immersion lens using a Nikon fluorescent microscope.
Note: Store the slides in the dark at 4 °C if necessary.
Data analysis
Microscope slides are imaged by a Nikon fluorescent microscope with a 60x water immersion lens. FITC and TRITC filter sets are used to visualize β-tubulin and PLA signals, respectively. The number of discrete fluorescent PLA spots inside the cells is counted. The average numbers of PLA signals per cell in different transfections are compared to investigate the change of interaction between the two fragments. The PLA signals from at least 60 cells are counted, and three independent experiments are performed. Statistical analysis is performed by using one-way analysis of variance (ANOVA) with Bonferroni post-hoc test.
Representative data
Recently, we demonstrated that the interaction of ELMO11-315 and ELMO1315-727, which contain its EID and EAD, respectively, in cells by PLA. Moreover, the number of PLA signals reduced significantly in cells co-transfected with the wildtype FE65 but not the binding defective mutant (i.e., FE65m) (Figure 4). These findings from the PLAs are in line with the result from the classical protein binding assay (Li et al., 2018).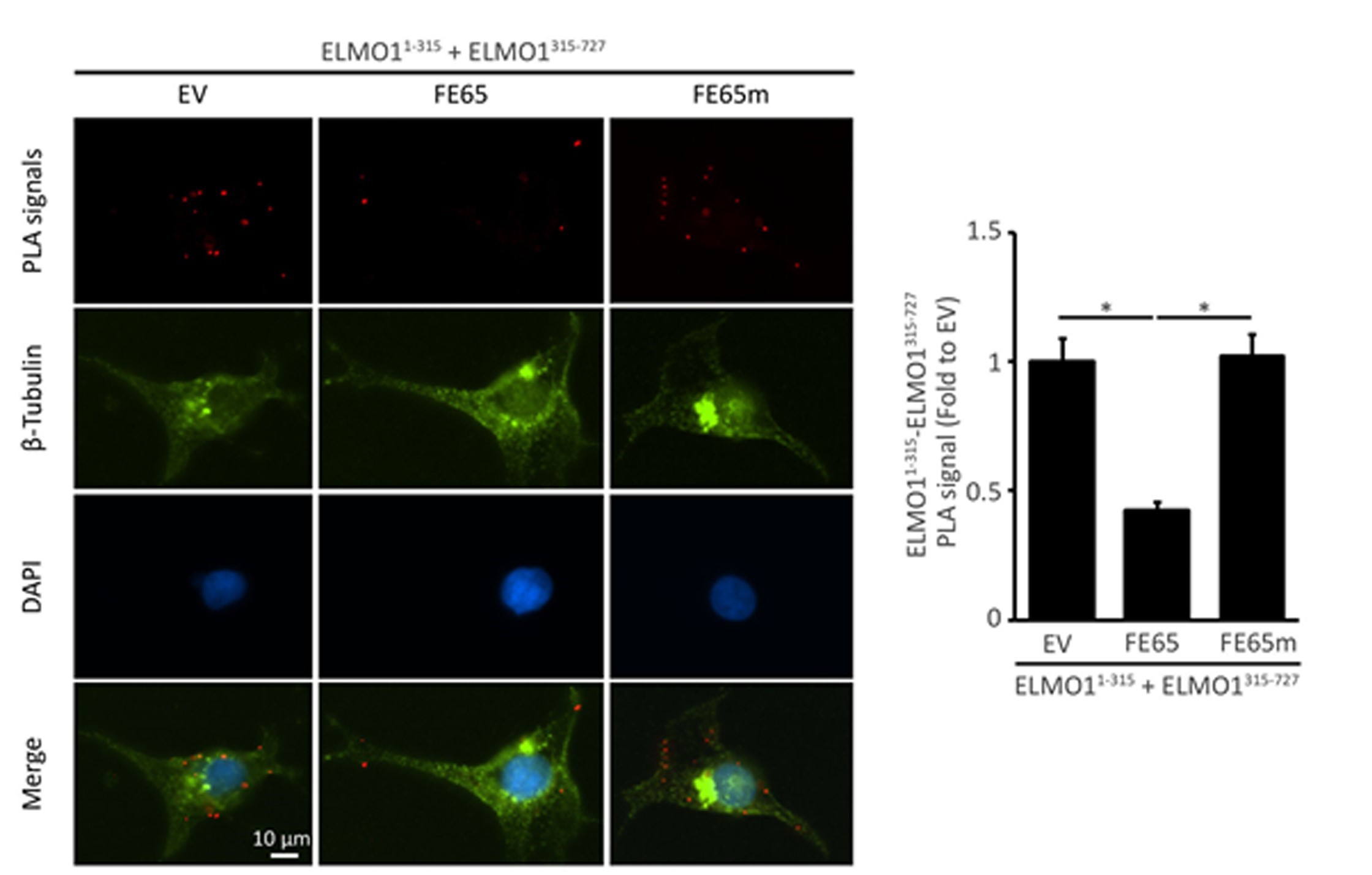
Figure 4. Representative images and quantification of PLA signals. HEK293 cells were transfected with myc-ELMO11-315 and FLAG-ELMO1315-727 together with either empty vector (EV), FE65 or FE65m. After 24 h transfection, the cells were fixed and subjected to PLA. The number of PLA signals from at least 60 cells was counted in each transfection and three independent experiments were performed. The bar chart shows the relative PLA signal (fold change to EV) in different transfections. *P < 0.001. Error bars are SEM. Significant reduction in PLA signals was observed only in FE65-transfected cells but not in EV- or FE65m-transfected cells. β-tubulin and nucleus were stained to serve as a morphological and nuclear marker, respectively. Scale bar = 10 μm. (This research was originally published in the Journal of Biological Chemistry. Li W, Tam KMV, Chan WWR, Koon AC, Ngo JCK, Chan HYE and Lau KF. Neuronal adaptor FE65 stimulates Rac1-mediated neurite outgrowth by recruiting and activating ELMO1. J. Biol. Chem. 2018; 293(20): 7674-7688. © the American Society for Biochemistry and Molecular Biology)
Recipes
- 1x TAE buffer
Dilute the 50x stock solution to 1x with distilled water - Poly-D-lysine solution
5 mg poly-D-lysine
5 ml distilled water
Dissolve 5 mg poly-D-lysine in 5 ml distilled water to obtain a stock solution (200x)
Store the stock solution in aliquots at -20 °C - 4% paraformaldehyde (freshly prepared)
0.4 g paraformaldehyde powder
10 ml PBS- Add 0.4 g paraformaldehyde powder to 10 ml PBS
- Incubate in a 70 °C water bath until the powder dissolved
- Cooldown the solution at room temperature
- Filter the solution with a 0.22 μm syringe filter
- 1x Ligation buffer
For each reaction, dilute 16 μl 5x Ligation buffer with 64 μl distilled water - 1x Amplification red
For each reaction, dilute 16 μl 5x Amplification Red with 64 μl distilled water - Wash buffer A
0.01 M Tris
0.15 M NaCl
0.05% Tween 20
pH 7.4 - Wash buffer B
0.2 M Tris
0.1 M NaCl
pH 7.5
Acknowledgments
This work was supported by funds from the Research Grants Council Hong Kong, Health and Medical Research Fund (Hong Kong), CUHK direct grant scheme and the United College endowment fund. This protocol was adapted from previous work. We thank Prof Jean-François Côté (Montreal Clinical Research Institute) for the mammalian expression construct of pcDNA3-N-Myc-Elmo1.
Competing interests
The authors declare that they have no conflicts of interest with the contents of this article.
References
- Li, W., Tam, K. M. V., Chan, W. W. R., Koon, A. C., Ngo, J. C. K., Chan, H. Y. E. and Lau, K. F. (2018). Neuronal adaptor FE65 stimulates Rac1-mediated neurite outgrowth by recruiting and activating ELMO1. J Biol Chem 293(20): 7674-7688.
- Miernyk, J. A. and Thelen, J. J. (2008). Biochemical approaches for discovering protein-protein interactions. Plant J 53(4): 597-609.
- Ohad, N., Shichrur, K. and Yalovsky, S. (2007). The analysis of protein-protein interactions in plants by bimolecular fluorescence complementation. Plant Physiol 145(4): 1090-1099.
- Patel, M., Margaron, Y., Fradet, N., Yang, Q., Wilkes, B., Bouvier, M., Hofmann, K. and Cote, J. F. (2010). An evolutionarily conserved autoinhibitory molecular switch in ELMO proteins regulates Rac signaling. Curr Biol 20(22): 2021-2027.
- Patel, M., Pelletier, A. and Cote, J. F. (2011). Opening up on ELMO regulation: New insights into the control of Rac signaling by the DOCK180/ELMO complex. Small GTPases 2(5): 268-275.
- Weibrecht, I., Leuchowius, K. J., Clausson, C. M., Conze, T., Jarvius, M., Howell, W. M., Kamali-Moghaddam, M. and Soderberg, O. (2010). Proximity ligation assays: a recent addition to the proteomics toolbox. Expert Rev Proteomics 7(3): 401-409.
Article Information
Copyright
© 2019 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Chan, W. W. R., Chau, D. L. D., Li, W. and Lau, K. (2019). Proximity Ligation Assay for the Investigation of the Intramolecular Interaction of ELMO1. Bio-protocol 9(23): e3449. DOI: 10.21769/BioProtoc.3449.
- Li, W., Tam, K. M. V., Chan, W. W. R., Koon, A. C., Ngo, J. C. K., Chan, H. Y. E. and Lau, K. F. (2018). Neuronal adaptor FE65 stimulates Rac1-mediated neurite outgrowth by recruiting and activating ELMO1. J Biol Chem 293(20): 7674-7688.
Category
Biochemistry > Protein > Interaction > Protein-protein interaction
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








