- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Bacterial Synchronized Transfer Assays in Bone Marrow Derived Macrophages
Published: Vol 9, Iss 22, Nov 20, 2019 DOI: 10.21769/BioProtoc.3437 Views: 5114
Reviewed by: Kristin L. ShinglerDarrell CockburnAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Quantification of Macrophage Cellular Ferrous Iron (Fe2+) Content using a Highly Specific Fluorescent Probe in a Plate-Reader
Philipp Grubwieser [...] Christa Pfeifhofer-Obermair
Feb 5, 2024 2215 Views

Quantitative Measurement of Plasma Membrane Protein Internalisation and Recycling in Heterogenous Cellular Samples by Flow Cytometry
Hui Jing Lim and Hamish E. G. McWilliam
May 5, 2024 2243 Views

Novel Experimental Approach to Investigate Immune Control of Vascular Function: Co-culture of Murine Aortas With T Lymphocytes or Macrophages
Taylor C. Kress [...] Eric J. Belin de Chantemèle
Sep 5, 2025 3547 Views
Abstract
Merocytophagy (“mero”, Greek for partial; “cytophagy” for cell eating) is a process by which cells acquire microbes and cytosolic material through phagocytosis of a small portion of neighboring cells upon cell-cell contact. Cell-cell contact dependent transfer events can be assessed through co-incubation of differently labeled cells. With these assays, it is difficult to analyze the recipient cells by microscopy or bacterial burden within only recipient cells. Therefore, we established a synchronized transfer assay that allows for recipient cells to be isolated from donor cells following transfer events at a high purity. Here, we present this assay in context of bacterial infections and cytosolic cellular staining. With this protocol, mechanisms of cell-cell contact dependent transfer events and the events following merocytophagy can easily be investigated.
Keywords: Cell transferBackground
Phagocytic cells can acquire infectious material when microbes are extracellular or directly from infected cells (Steele et al., 2016). This host mediated process allows for initiation of an effective cellular response to clear infection (Ahmed et al., 2008). However, this process can be exploited by bacterial pathogens to expand their replicative niche (Ireton, 2013; Steele et al., 2016). We previously found that cells can acquire microbes and cytosolic material from neighboring cells through phagocytosis of a small portion of the infected cell which does not kill the donor cell (Steele et al., 2016 and 2019). This process known as merocytophagy (“mero”, Greek for partial; “cytophagy” for cell eating) provides a mechanism for the intracellular bacteria to disseminate and sustain infection (Steele et al., 2016 and 2019).
To understand the mechanism of merocytophagy, we established a synchronized transfer assay that can be used to analyze a purified population of recipient cells following transfer events. Previously described transfer assays allow for the quantification of transfer events, whereas this assay allows for the analysis of the engulfed material. Specifically, engulfed material following a transfer event can be analyzed using microscopy or microbial burdens. Herein, we describe a detailed protocol of the synchronized transfer assay to analyze transferred bacteria and cytosolic dye.
Materials and Reagents
- Serological Pipette (e.g., Thermo Scientific, catalog number: 14-387-163)
- 250 ml filter unit, 0.2 micron PES membrane filter, sterile (VWR, catalog number: 10040-464)
- 10 ml syringes (Fisher Scientific, catalog number: 201635)
- FisherbrandTM Easy ReaderTM Conical Polypropylene centrifuge tube 15 ml (Fisher Scientific, catalog number: 05-539-12)
- FisherbrandTM Easy ReaderTM Conical Polypropylene centrifuge tube 50 ml (Fisher Scientific, catalog number: 05-539-13)
- FisherbrandTM Microscope Slides, Frosted (Fisher Scientific, catalog number: 12-552-3)
- FisherbrandTM Non-Tissue Culture treated Petri Dishes with Clear Lid (Fisher Scientific, catalog number: FB0875712)
- Serological Pipette Tips (Various Sizes) (Fisher Scientific, catalog numbers: 07-200-571, 07-200-574, 07-200-573, 07-200-575)
- FisherbrandTM Cell Strainer (40 μM) (Fisher Scientific, catalog number: 22363547)
- Matsunami Micro Cover Glasses, Round (18 mm diameter) (VWRTM, catalog number: 10026-138)
- Micropipette Tips (Various Sizes) (VWR, catalog numbers: 10017-062, 10017-064, 10017-66, 1007-068, 1007-092)
- Corning Costar Flat Bottom Cell Culture Plates (Fisher Scientific, catalog number: 3513)
- 26G BD Precision Glide needles (Fisher Scientific, catalog number: 148266A)
- C57BL/6J Mice (Jackson Laboratory, Stock No. 000664)
- Fetal Bovine Serum (FBS) (Atlanta Biologicals, catalog number: S10350)
- Francisella tularensis subsp. holarctica inducible T6SS mutant (Steele et al., 2019)
- Francisella tularensis subsp. holarctica live vaccine strain (LVS) (Center for Disease Control, CDC)
- Recombinant murine M-CSF (rmM-CSF) (Peprotech, catalog number: 315-02)
- RemelTM Hemoglobin (Fisher Scientific, catalog number: R451402)
- J774A-1 Macrophages (ATCC® TIB-67)
- L929 fibroblasts (ATCC® CLL-1)
- GibcoTM TrypLE Express Enzyme (1x) (Fisher Scientific, catalog number: 12-605-028)
- L-Aspartic Acid (Arcos Organics, catalog number: 56-84-8)
- DL-Isoleucine (Fisher Scientific, catalog number: A17521)
- L-Leucine (Arcos Organics, catalog number: 12512-100)
- DL-Methionine (Arcos Organics, catalog number: 59-51-8)
- DL-Serine (Fisher Scientific, catalog number: A15184)
- L-Tyrosine (Fisher Scientific, catalog number: J63511)
- L-Cysteine (Sigma-Aldrich, catalog number C7602-25G)
- Thiamine HCl (Arcos Organics, catalog number: 67-03-8)
- Spermine HCl (Fisher Scientific, catalog number: 5677-5GM)
- L-Arginine (Arcos Organics, catalog number: 74-79-3)
- L-Histidine (Arcos Organics, catalog number: 71-00-1)
- L-Lysine (Fisher Scientific, catalog number: 657-27-2)
- L-Proline (Fisher Scientific, catalog number: A10199)
- DL-Threonine (Fisher Scientific, catalog number: A10606)
- DL-Valine (Arcos Organics, catalog number: 516-06-3)
- Ammonium Chloride (NH4Cl) (ACROS Organics, catalog number: AC423285000)
- 4% Paraformaldehyde (PFA) (Fisher Scientific, catalog number: AAJ19943K2)
- Bacto Agar (Fisher Scientific, catalog number: DF1040010)
- Brain Heart Infusions (BHI, Fisher Scientific, catalog number: DF0037178)
- Cell Trace Far Red (CTR) Dye (ThermoFisher, catalog number: C34564)
- Cell Trace CFSE (CFSE) (ThermoFisher, catalog number: C34553)
- CorningTM RPMI 1640 Medium (Mod.) 1x with L-Glutamine (Fisher Scientific, catalog number: MT10-040-CV)
- Dulbecco’s Modification of Eagles Medium (DMEM) with 4.5 g/L glucose, L-glutamine, sodium pyruvate (Corning Life Sciences, catalog number: 10-013-CM)
- DPBS (with magnesium (Mg) and calcium (Ca) (Corning Life Sciences, catalog number: 21-030-CM)
- DPBS (without magnesium (Mg) and calcium (Ca) (Corning Life Sciences, catalog number: 21-031-CM)
- EDTA disodium salt (ACROS Organics, catalog number: AC409975000)
- GibcoTM GlutaMAXTM Supplement (Fisher Scientific, catalog number: 35050-061)
- GibcoTM Sodium Bicarbonate 7.5% solution (Fisher Scientific, catalog number: 25080-094)
- Gibco Penicillin-Streptomycin (10,000 U/ml) (Fisher Scientific, catalog number: 15140-148)
- HyCloneTM MEM Non-essential Amino acids (Fisher Scientific, catalog number: SH30238.01)
- HyCloneTM Sodium pyruvate (Fisher Scientific, catalog number: SH30239.01)
- Potassium Bicarbonate (KHCO3) (Fisher Scientific, catalog number: P184-500)
- Sodium Hydroxide (Fisher Scientific, catalog number: SS255-1)
- Trypan Blue (MP Biomedicals, catalog number: 195532)
- Vectashield Antifade Mounting Medium with DAPI (Vector Laboratories, catalog number H-1200)
- Calcium Pantothenate (Arcos Organics, catalog number: 137-08-6)
- Magnesium Sulfate (MgSO4·7H2O) (Arcos Organics, catalog number: 10034-99-8)
- Iron (II) Sulfate Heptahydrate (FeSO4·7H2O) (Fisher Scientific, catalog number: A15178)
- Potassium Phosphate Monobasic (KH2PO4) (Fisher Scientific, catalog number: P285-500)
- Potassium Phosphate Dibasic Anhydrous (K2HPO4) (Fisher Scientific, catalog number: P288-100)
- D-Glucose (Fisher Scientific, catalog number: A16828)
- HCl
- NaCl
- Isovitalex
- 10% FBS DMEM (see Recipes)
- ACK lysis buffer (see Recipes)
- Chocolate agar plates (see Recipes)
- Chamberlain’s defined medium (CDM) (Chamberlain et al. [1965]) (see Recipes)
- L cell media (see Recipes)
- Bone marrow differentiation media (see Recipes)
- 0.5 M EDTA (see Recipes)
- 10 mM EDTA in 1x DPBS (without Mg and Ca) (see Recipes)
- 10x Glucose Salts (see Recipes)
Equipment
- Blunt forceps (Fisher Scientific, catalog number: 08-875-8B)
- T75 flasks (Fisher Scientific, catalog number: 130190)
- T175 flasks (Fisher Scientific, catalog number: 130191)
- Surgical Scissors (Fisher Scientific, catalog number: 08-951-5)
- Micropipettes (e.g., Rainin, catalog numbers: 17008655, 17008650, 17008652, 17008653)
- Benchtop Swing Bucket Centrifuge (e.g., Eppendorf, catalog number: 022625004)
- Hemocytometer (e.g., Fisher Scientific, catalog number 02-671-6)
- Shaking Water bath (e.g., Fisher Scientific, catalog number: S37368)
- Humidified 5% CO2 incubators set to 37 °C (e.g., NuAire, NU-5700)
- Leica DM4000 upright fluorescence microscope
- OD spectrometer
- Light Microscope
- Biosafety cabinet
Software
- Leica Application Suite X (LAS X) (Leica microsystems, www.leica-microsystems.com), for microscopy analysis
Procedure
- L929 cell conditional media preparation
- Grow L929 fibroblasts to confluency in two T75 flasks using L cell media (Recipe 9). Incubate at 37 °C 5% CO2 for approximately 2 days.
- Detach cells by removing existing media and adding 4 ml of TrypLE to flask.
- Incubate at 37 °C 5% CO2 for 5 mins or until cells are completely detached.
- Collect detached cells by adding 10 ml of L cells media to flask and transfer to 50 ml conical tube.
- Centrifuge at room temperature at 250-300 x g for 5 min to remove TrypLE from media. Re-suspend cells in L cell media.
- Count cells by adding 10 μl of trypan blue to 10 μl of cells. Add 10 μl of mixture to hemocytometer and count under light microscope.
- Plate cells at a density of 2.5 x 105 cells in 75 ml L cell media in T225 flasks.
- Rock flasks back and forth to get equal coverage of the entire flask bottom.
- Incubate at 37 °C 5% CO2 for 10-12 days (watch for cobblestone appearance of cells).
- Remove media from all of the flasks and place into 50 ml conical tubes.
- Centrifuge at 300 x g for 5 min at 4 °C.
- To ensure thorough mixing of all collected media, combine all of the supernatants into one 50 ml conical tube.
- Vacuum filter media through a 250 ml filter unit.
- Aliquot 45 ml into 50 ml conical tubes.
- Transfer tubes to -20 °C for long-term storage. Thaw media at 37 °C when needed for bone marrow macrophage differentiation.
- Bone marrow derived macrophages preparation
Note: Protocol can be performed using J774A.1 macrophage (ATCC® TIB-67), if bone marrow derived macrophages are unavailable. If performing EEA-1 staining, do not allow cells to fix for more than 7 min or antibody will not stain well.- Euthanize mouse by CO2 and cervical dislocation in accordance with IACUC protocol.
- Remove femurs from mouse in a biosafety cabinet.
Note: Keep bones intact and clean bones of any excess tissue. - For each femur, cut each end using sterile scissors. Cut approximately 1 mm off of each head of the bone or until you see an opening large enough for a 26G needle.
- With forceps, hold the femur over a 50 ml conical tube.
- Prepare a 10 ml syringe with needle. Fill syringe with 10 ml of DMEM.
- Insert needle into the opening of the bone.
- Flush femur with 5 ml of DMEM into the 50 ml conical tube.
Note: Be sure to stick the needle into each opening of the femur. If needle cannot enter the femur, cut more off of the end. - Repeat Steps B6 and B7 with second femur.
- Filter the solution with a 40 μm cell strainer into a fresh 50 ml conical tube to remove clumps of tissue.
- Centrifuge cells at room temperature at 250-300 x g for 5 min.
- Remove media and gently resuspend pellet in residual media (approximately 1 ml).
- Add 1 ml of ACK lysis buffer (Recipe 2) and incubate for 60 s.
- Add 9 ml of DMEM.
- Centrifuge cells at room temperature at 250-300 x g for 5 min.
- Remove media and resuspend cells in approximately 10 ml of L cell differentiation media (Recipe 1 and Procedure A).
- Seed approximately 5 ml in 20 ml final volume of media into non-tissue culture treated Petri dish.
- Allow cells to differentiate in 37 °C 5% CO2 incubator for 6 days.
Note: Ensure proper differentiation by checking morphology periodically. See Figure 1 for reference of proper differentiation (cells appear with elongated shape).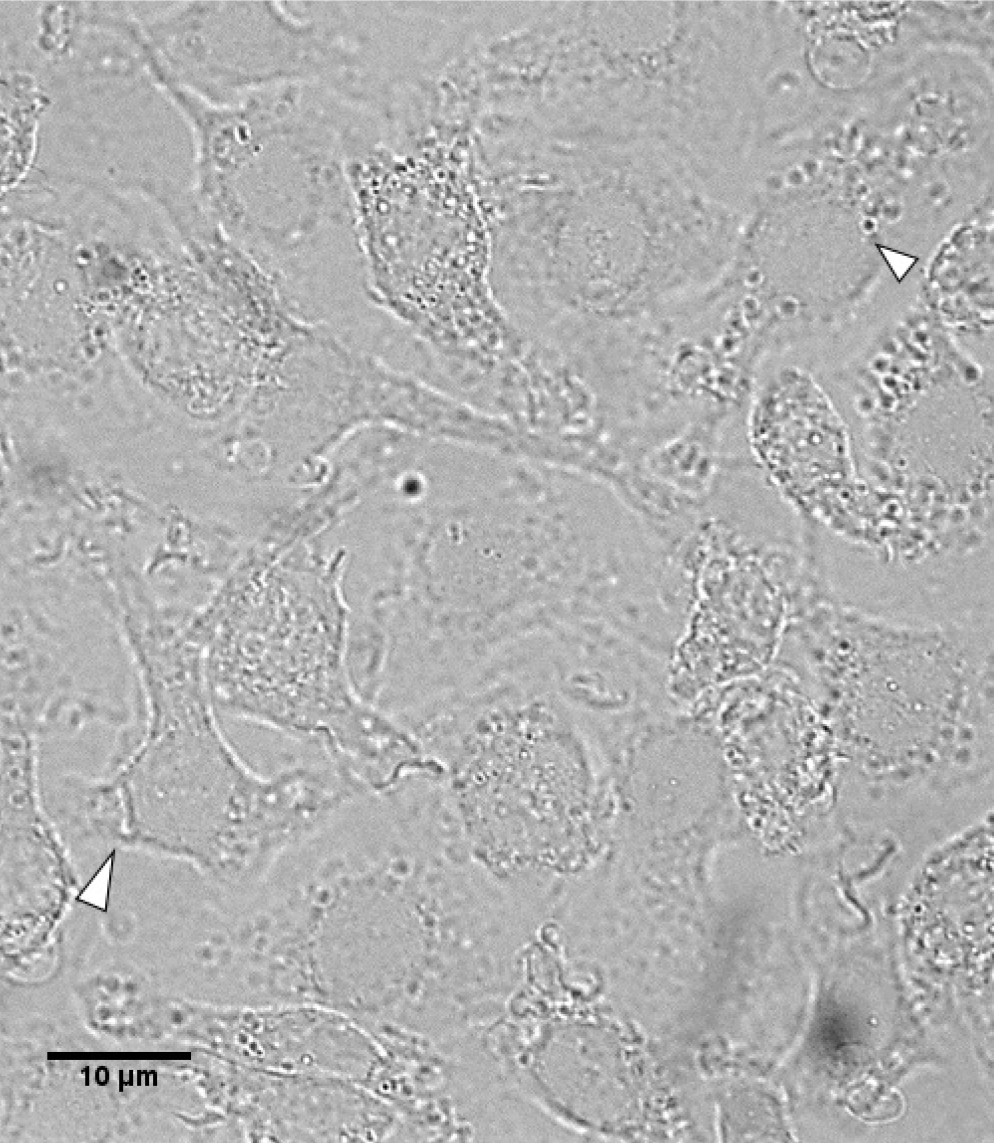
Figure 1. Morphology of Day 5 differentiated BMDMs. Appearance of elongated shape as indicated by arrowheads. - On Day 6, remove media from cells and wash cells by pipetting 10 ml of 1x DPBS (with Mg and Ca). Repeat wash 2 more times.
- Following final wash, add 3-4 ml/plate of 10 mM EDTA in 1x DPBS (without Mg and Ca), pH 7.2-7.4.
- Incubate plates at 37 °C for approximately 5 min or until cells begin to lift off.
- Add 5-8 ml/plate of DMEM with 10% FBS DMEM (Recipe 5) and transfer to a 15 ml conical tube.
- Centrifuge cells at room temperature at 250-300 x g for 5 min.
- Remove media and resuspend cells in 10 ml of DMEM with 10% FBS.
- Count cells using a hemocytometer.
Note: Expect a cell count of 6 to 8 million cells per plate. - Seed a 12-well tissue culture treated plate at 2 x 105 cells per well in a total volume of 1 ml per well.
Note: For each sample, seed 1 well with a coverslip (recipient sample) and 1 well without a coverslip (donor cells). - Incubate overnight at 37 °C 5% CO2.
- Transfer of cell permeable dye
Note: Wells should be set up with CFSE and CTR cells only (i.e. cells that do not have contact).- Stain donor cells with Cell Trace CFSE (CFSE) at a final concentration of 5 μM in dPBS with Mg and Ca (500 μl per well) in a 12-well plate.
- Stain recipient cells with Cell Trace Far Red (CTR) at a final concentration of 1 μM in dPBS with Mg and Ca (500 μl per well) in a 12-well plate.
- Incubate cells at 37 °C 5% CO2 for 20 min.
- Wash cells 3 times each with 1 ml of pre-warmed 10% FBS DMEM.
Note: 10% FBS DMEM should be pre-warmed in a 37 °C water bath for 20 min. - After the final wash, pick up the coverslip with recipient cells and invert it onto the well with donor cells so that the cells are in direct contact.
- Incubate for 30 min at 37 °C 5% CO2.
- Remove the coverslip and return it into the original well in its original orientation.
- At desired time point, add 4% PFA to coverslip (1 ml per well) and incubate at room temperature for 7 min (for EEA-1 staining) or 10 min for other antibodies.
- Wash coverslips 3 times with 1 ml of 1x DPBS (with Mg and Ca).
- Perform desired microscopy staining (e.g., LAMP-1 [Steele et al., 2019]).
- Wash coverslips 3 times with 1 ml of 1x DPBS.
- Mount coverslips onto slides using mounting media by adding approximately 10 μl of mounting media to slides and inverting coverslip onto slide.
- Bacterial infection
Note: Various bacteria can be used for this assay with modifications needed for growth and infection conditions. Control wells should be set up with CFSE and CTR cells only.- Streak out Francisella tularensis freezer stock on chocolate agar.
- Incubate plate at 37 °C for 3 days.
- Pick 4-6 individual colonies from the plate and suspend colonies in 3 ml of CDM.
- Incubate culture overnight at 37 °C in a shaking water bath.
- Next day, spin down 2 ml of culture at room temperature at 20,000 x g for 2 min.
- Remove the supernatant and resuspend each pellet in 1 ml of PBS.
- Blank OD spec with 1 ml of PBS.
- Measure OD600 of culture (desired OD should be 0.34-0.36, if not incubation should continue).
- Infect donor cells at MOI 100 in 250 μl total volume of 10% FBS.
- Incubate for 2 h at 37 °C 5% CO2.
- Add 10 μg/ml of gentamicin in a total volume of 500 μl per well.
- Twenty hours following inoculation of bacteria, replace media on all samples.
- Invert coverslip of recipient cells onto donor cells to allow for direct contact.
- Incubate for 30 min at 37 °C.
- Remove the coverslip and put it into the original well in its original orientation.
- At desired time point, add 4% PFA to coverslip (1 ml per well) and incubate at room temperature for 7 min (for EEA-1 staining) or 10 min for other antibodies.
- Wash coverslips 3 times with 1 ml of 1x DPBS (with Mg and Ca).
- Perform desired microscopy staining (e.g., LAMP-1 [Steele et al., 2019])
- Wash coverslips 3 times with 1 ml of 1x DPBS.
- Mount coverslips onto slides using mounting media by adding approximately 10 uμl of mounting media to slides and inverting coverslip onto slide.
Data analysis
Microscopy results were collected using a Leica DM400 upright fluorescence microscope. For each sample, 50 images were taken at each desired time point per experiment. Images were analyzed using Leica LAS X software. The number of cells that acquired material from donor cells can be determined by counting the number of recipient cells that obtained either bacteria or CFSE (Figure 2 for reference). Transwells (or physical separation) can be used to determine the number of phagocytosis events of extracellular material (Steele et al., 2016 and 2019).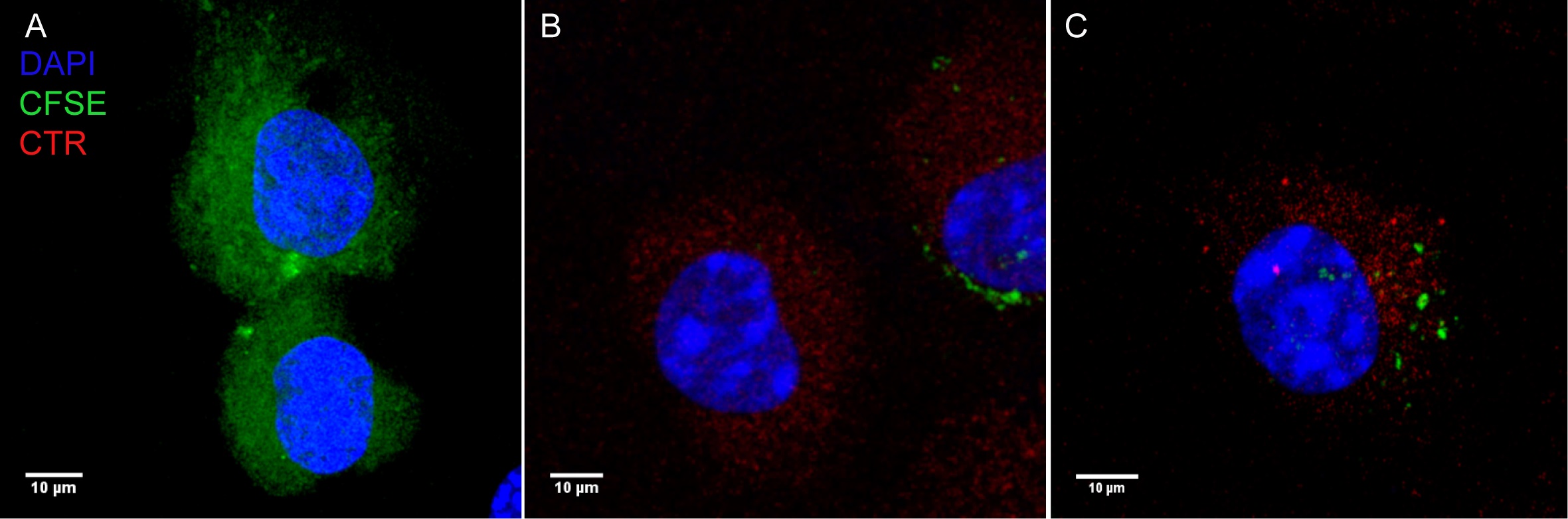
Figure 2. Representative microscopy of CFSE transfer events. A. CFSE stained donor BMDMs. B. CTR stained recipient BMDM. C. CFSE transfer to CTR stained recipient BMDM.
Recipes
- Bone marrow differentiation media (150 ml for 1 mouse)
75 ml of DMEM
30 ml of heat inactivated FBS
45 ml L929 conditional media (can be substituted with M-CSF, Griffin et al., 2013)
1.5 ml of 100 mM L-glutamine (or GlutaMax)
1.5 ml of 100 mM sodium pyruvate
1.5 ml of 7.5% sodium bicarbonate solution
Mix and filter sterilize - ACK lysis buffer
8.3 g of NH4Cl
1.0 g of KHCO3
200 μl of 0.5 M EDTA
Add ddH2O to 1 L, adjust to pH 7.2-7.4 and filter sterilize - 0.5 M EDTA
Dissolve 186.1 g EDTA disodium salt in 750 ml of ddH2O
Add 15 g solid NaOH to solution to solubilize EDTA
Use 5 N NaOH to adjust pH to 7.5-8.0 - 10 mM EDTA in 1x DPBS (without Mg and Ca)
Add 20 ml of 0.5 M EDTA to 1 L of 1x DPBS (without Mg and Ca) - 10% FBS DMEM
Add 100 ml of thawed heat inactivated FBS to 1 L of DMEM - Chamberlain’s defined medium (CDM) (Chamberlain et al., 1965)
Add components to a final concentration of 1x in a desired volume of H2O:
L-Aspartic Acid (100x stock: 2 g into 50 ml HCl)
DL-Isoleucine (100x stock: 2 g into 50 ml HCl)
L-Leucine (100x stock: 2 g into 50 ml HCl)
DL-Methionine (100x stock: 2 g into 50 ml HCl)
DL-Serine (100x stock: 2 g into 50 ml HCl)
L-Tyrosine (100x stock: 2 g into 50 ml HCl)
L-Cysteine (100x stock: 1 g into 50 ml HCl)
Thiamine HCl (100x stock: 200 mg into 50 ml HCl)
Spermine HCl (100x stock: 200 mg into 50 ml HCl)
L-Arginine (100x stock: 2 g into 50 ml H2O)
L-Histidine (100x stock: 1 g into 50 ml H2O)
L-Lysine (100x stock: 2 g into 50 ml H2O)
L-Proline (100x stock: 5 g into 50 ml H2O)
DL-Threonine (100x stock: 2.5 g into 50 ml H2O)
DL-Valine (100x stock: 2 g into 50 ml H2O)
Calcium Pantothenate (100x stock: 100 mg into 50 ml H2O)
10x Glucose Salts (see Recipe 7)
Once all components are added, pH solution to 6.2-6.4 - 10x Glucose salts
25 g NaCl
338 mg MgSO4·7H2O
5 mg FeSO4·7H2O
2.5 g KH2PO4
2.5 g K2HPO4
10 g Glucose
250 ml H2O - Chocolate agar plates
In a 2 L flask, add:
25 g of BHI
15 g of Bacto Agar
500 ml of H2O
In a second 2 L flask add:
10 g of hemoglobin
500 ml of H2O
Autoclave for 20 min on liquid cycle
Remove media from autoclave and place into a 50 °C water bath
Once cooled, add 10 ml of isovitalex
Combine hemoglobin to BHI and gently stir by hand - L cell media
500 ml of RPMI 1640
50 ml of heat inactivated FBS
2.5 ml of Pen/strep
5 ml of 100 mM sodium pyruvate
5 ml 100 mM non-essential amino acids
Acknowledgments
The authors would like to thank the National Institute of Allergy and Infectious Diseases (NIH) for funding (R01AI082870 and R56AI139476).
Competing interests
No competing interests declared.
Ethics
All animals were handled according to approved institutional animal care and use committee (IACUC) protocol #4946 at Washington State University.
References
- Ahmed, K. A., Munegowda, M. A., Xie, Y. and Xiang, J. (2008). Intercellular trogocytosis plays an important role in modulation of immune responses. Cell Mol Immunol 5(4): 261-269.
- Chamberlain, R. E. (1965). Evaluation of live tularemia vaccine prepared in a chemically defined medium. Appl Microbiol 13: 232-235.
- Griffin, A. J., Crane, D. D., Wehrly, T. D., Scott, D. P. and Bosio, C. M. (2013). Alternative activation of macrophages and induction of arginase are not components of pathogenesis mediated by Francisella species. PLoS One 8(12): e82096.
- Ireton, K. (2013). Molecular mechanisms of cell-cell spread of intracellular bacterial pathogens. Open Biol 3(7): 130079.
- Steele, S. P., Chamberlain, Z., Park, J. and Kawula, T. H. (2019). Francisella tularensis enters a double membraned compartment following cell-cell transfer. Elife 8: e45252.
- Steele, S., Radlinski, L., Taft-Benz, S., Brunton, J. and Kawula, T. H. (2016). Trogocytosis-associated cell to cell spread of intracellular bacterial pathogens. Elife 5: e10625.
Article Information
Copyright
Dominguez et al. This article is distributed under the terms of the Creative Commons Attribution License (CC BY 4.0).
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Dominguez, S. R., Kawula, T. and Steele, S. P. (2019). Bacterial Synchronized Transfer Assays in Bone Marrow Derived Macrophages. Bio-protocol 9(22): e3437. DOI: 10.21769/BioProtoc.3437.
- Steele, S. P., Chamberlain, Z., Park, J. and Kawula, T. H. (2019). Francisella tularensis enters a double membraned compartment following cell-cell transfer. Elife 8: e45252.
Category
Immunology > Immune cell function > Macrophage
Cell Biology > Cell-based analysis > Endocytosis
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









