- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Leishmania Parasite Quantification by Bioluminescence in Murine Models
Published: Vol 9, Iss 22, Nov 20, 2019 DOI: 10.21769/BioProtoc.3431 Views: 4723
Reviewed by: Alexandros AlexandratosMahmoud Kamal AhmadiOlga Koutsoni

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Optimizing Confocal Imaging Protocols for Muscle Fiber Typing in the Mouse Masseter Muscle
Catalina Matias [...] Jeffrey J. Brault
Apr 5, 2025 2950 Views

Development of a Novel Automated Workflow in Fiji ImageJ for Batch Analysis of Confocal Imaging Data to Quantify Protein Colocalization Using Manders Coefficient
Vikram Aditya [...] Wei Yue
Apr 5, 2025 2930 Views

Detecting the Activation of Endogenous Small GTPases via Fluorescent Signals Utilizing a Split mNeonGreen: Small GTPase ActIvitY ANalyzing (SAIYAN) System
Miharu Maeda and Kota Saito
Jan 5, 2026 525 Views
Abstract
Leishmaniasis remains a major public health problem worldwide with a prevalence of 12 million, an incidence of 1 million persons, and 350 million people being at risk. Murine models have been largely used for studying the host-pathogen relationship and developing effective chemotherapies against Leishmania parasites. Thus, preclinical imaging is crucial for monitoring the disease outcome. The aim of this protocol is to quantify parasite burden using bioluminescence in vivo imaging. Here, we describe a high-throughput imaging workflow, together with data acquisition and analysis ideal to assess in vivo parasite load in mouse models.
Keywords: ParasiteBackground
Leishmania parasites are the causative agents of the neglected tropical disease known as leishmaniasis. The disease outcome varies depending on different factors including the infecting species, the immune response of the host, the presence of an endosymbiotic virus within Leishmania parasites (Ives et al., 2011), or the co-infection with viruses such human immunodeficiency virus (HIV) or lymphocytic choriomeningitis mammarenavirus (LCMV) which can be detrimental for disease exacerbation (van Griensven et al., 2014; Rossi et al., 2017). Therefore, the study of parasite persistence using murine models has emerged in the past years (Eren et al., 2016). The quantification of parasite burden in experimental models of Leishmania infection has been commonly quantified by either Real-Time Quantitative Reverse Transcription PCR (qRT-PCR) or by limiting dilution assays upon euthanasia of animals (Hartley et al., 2016). In this protocol, we report a non-invasive technique for the quantification of parasite burden during the disease progression using luciferase expressing parasites (Ives et al., 2011; Hartley et al., 2018). Moreover, this approach can also be applied for the in vivo detection of other chemiluminescent agents such as luminol that is used as a readout of myeloperoxidase (MPO) activity in acute and chronic inflammatory diseases, or for any other organism expressing luciferase.
Materials and Reagents
- Eco Nitrile PF 250 Gloves (ecoSHIELDTM)
- Tissue culture flask 25 version “Vent” (TPP®, catalog number: 90025)
- Microcentrifuge 1.5 ml tubes (Corning, Axygen®, catalog number: MCT-175-C)
- 50 ml syringe (B. Braun Medical, catalog number: 4617509F-02)
- Polypropylene conical 15 ml centrifuge tube (TPP Techno Plastic Products, catalog number: 91015)
- Polypropylene conical 50 ml centrifuge tube (TPP Techno Plastic Products, catalog number: 91050)
- BD Micro-FineTM + U-100 Insulin syringes 0.5 ml 0.33 mm (29G) x 12.7 mm (BD Medical, catalog number: 324824)
- 2 ml serological pipettes (FALCON®, catalog number: 357507)
- 10 ml serological pipettes (SARSTEDT, catalog number: 86.1254. 001)
- 25 ml serological pipettes (SARSTEDT, catalog number: 86.1685.001
- 0.22 μm syringe-filter (Carl Roth, catalog number: P668.1)
- Vacuum Filtration 500 “rapid”-Filtermax, PES membrane 0.22 μm pore size (TPP®, catalog number: 99500)
- 6-9-week-old specific-pathogen free C57BL/6 female mic
- Dulbecco's Phosphate-Buffered Saline (DPBS) (Thermo Fisher Scientific, GibcoTM, catalog number: 14040091), stored at 4 °C
- Schneider’s Drosophila Medium w: L-Glutamine and 0.40 g/L NaHCO3 (PANTM BIOTECH, catalog number: P04-91500), store at 4 °C
- Fetal Bovine Serum (FBS) (Thermo Fisher Scientific, GibcoTM, catalog number: 10270106), stored at -20 °C
- HEPES Buffer (1M) (BioConcept, catalog Number: 5-31F00-H), stored at 4 °C
- Penicillin-Streptomycin (P/S) solution (10.000 IU/ml P/ 10 mg/ml S) (Bioconcept, catalog number: 4-01F00-H), stored at 4 °C
- Hemin BioXtra, from Porcine (Sigma-Aldrich, catalog number: 51280), stored at 4 °C
- Folic acid (FlukaTM, catalog number: 47620), stored at 4 °C
- 6-Biopterin (Sigma-Aldrich, catalog number: B2517), stored at -20 °C
- Dimethyl sulfoxide (DMSO) Cell culture grade (AppliChem GmbH, catalog number: A3672)
- Sodium hydroxide ≥ 98% p.a., ISO, in pellets (Carl Roth, catalog number: 6771.1)
- Glutaraldehyde solution technical, ~50% in H2O (5.6 M) (FlukaTM, catalog number: 49629), stored at 4 °C
- 70% Ethanol F25® (in H2O) (Alcosuisse)
- VivoGloTM Luciferin, in vivo Grade (Promega, catalog number: P1041), stored at -20 °C
- Luciferase-expressing transfectants of Leishmania guyanensis parasites M4147 (MHOM/BR/75/M4147)
- AttaneTM Isoflurane ad us. vet. (Provet AG, catalog number: covetrus 722 2222)
- Complete parasite culture medium (see Recipes), stored at 4 °C
- Heat-inactivated FBS (see Recipes)
- Biopterin (see Recipes)
- Hemin-folate (see Recipes)
- Parasite fixative solution (see Recipes), stored at 4 °C
- Luciferin solution (see Recipes), stored at -20 °C
Equipment
- FormaTM Steri-CycleTM CO2 Incubator at 26 °C (Thermo Fisher Scientific, catalog number: 371)
- Pipette controller (INTEGRA, catalog number: 155017)
- Pipettes (Gilson®)
- Counting chamber improved Neubauer (Assistent®, catalog number: 40442)
- Centrifuge 5810 R (Eppendorf)
- Benchtop pH Meter (METTLER TOLEDO)
- Water Bath (Julabo)
- Laminar flow hood parasite culture (SafeFAST Premium)
- Laminar flow hood mouse handling (Scanlaf Safe 1200)
- Combi-vet Anesthesia machine with 4 Flowmeters (Rothacher Medical GmbH)
- O2 Concentrator MIC 02 10 lpm for Anesthesia system (Rothacher Medical GmbH, catalog number: CV 10-101 MIC 10 lpm)
- Anesthetic waste gas filter (Aldasorber 1400g)
- Veterinary Anesthesia Induction chamber/for rodents (Rothacher Medical GmbH, catalog number: PS-0346)
- In vivo Imaging System (In-Vivo Xtreme II, BRUKER)
- In-Vivo XtremeTM II SPF animal chamber (SPFC) with HEPA filters (BRUKER)
- Inverted microscope (Olympus CKX41)
- Inverted confocal microscope (Zeiss LSM 800)
- Computer (Windows 7)
Software
- Molecular Imaging (MI) software version v. 7.5.3.22464 (BRUKER)
- GraphPad Prism software version 8.1.0 (GraphPad)
Procedure
- Preparation of luciferase-expressing transfectants of Leishmania guyanensis (Lgy) parasites
Note: The parasites are derived from the parental strain Lgy M4147 (MHOM/BR/75/M4147) and express firefly luciferase (5 x 107 photons/sec/106 parasites) from a LUC gene integrated stably into the small subunit of the ribosomal RNA locus (Lye et al., 2010). Stock parasites are maintained in liquid nitrogen. When starting from a frozen vial, thaw the parasites quickly by immersing the vial in a water bath at 26 °C. Transfer the parasites to a 15 ml tube and add 5 ml of Schneider medium (incomplete), resuspend and centrifuge at 2000 x g for 8 min at 26 °C. Remove the supernatant, resuspend in 5 ml of complete parasite culture medium and transfer into a 25 cm2 flask. Incubate at 26 °C and 5% CO2. The parasites were passaged in culture for not more than five passages. The parasites are isolated from mouse footpads to maintain their virulence and avoiding purification of metacyclic promastigotes and thus diminishing the risk of LPS contamination which could occur during purification and would activate the innate immune response and possibly affect read-outs.- Dilute 100 μl of parasites (around 5 x 106 parasites) in 10 ml of complete parasite culture medium using 25 cm2 flask (Recipe 1).
- Incubate the parasites for 6 days at 26 °C and 5% CO2.
- At Day 6 of culture, verify that the parasites are in stationary growth phase (Figure 1).
Note: Parasites are counted every day, once they reach their stationary phase, they are used for mice infection. - Count the parasites:
- Remove 10 μl of the parasite culture and transfer it to a 1.5 ml microcentrifuge tube containing 990 μl of parasite fixative solution (Recipe 5).
- Wait for 3 min.
- Take 10 μl and place in the counting chamber.
- Count the parasites using an inverted microscope with 20x objective.
- Adjust parasite number to 20 x 106/ml in DPBS.
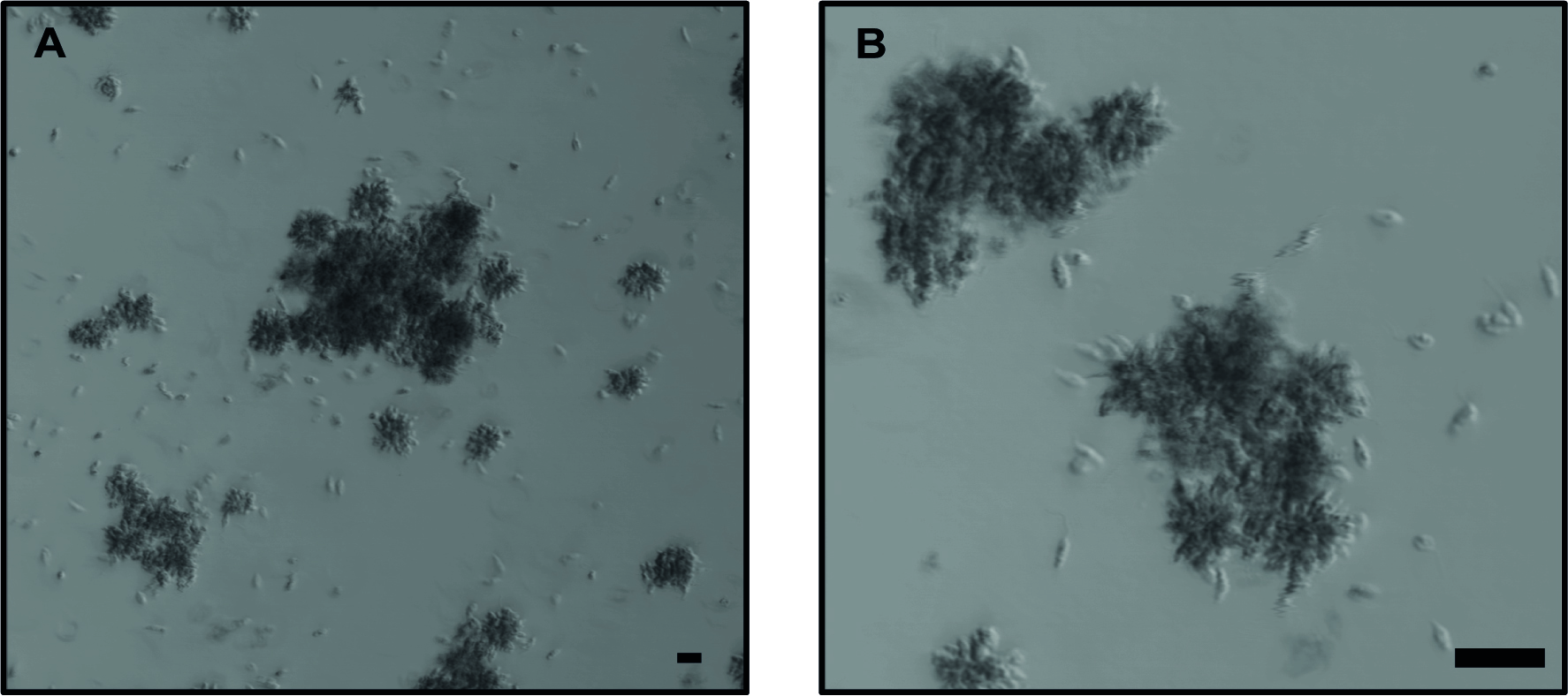
Figure 1. Lgy parasites after 6 days in culture. Representative images of parasites in stationary phase forming large clumps, also known as “rosettes”. A. 10x objective without zoom. B. 10x objective with 4x zoom. Bright-field images using confocal microscope. The scale bar represents 20 μm.
- Mice infection with Lgy parasites
Notes:- All mice handling steps are performed in clean conditions under a laminar flow hood. We use C57BL/6 because they show a healing phenotype and therefore less gravity on the mice and even with a darker tissue, light transmission can be measured (Ives et al., 2011).
- Parasites are directly inoculated into the mice after adjusting to 20 x 106 parasites/ml. Injection of parasites can be performed in one or both footpads without any difference.
- Slightly anesthetize the mice:
- Fill the isoflurane tank (Video 1, part 1).
- Switch on the O2 concentrator.
- Set the adjusting knob of the Combi-vet Anesthesia machine at 3% of isoflurane concentration.
- Switch on the flowmeter connected to the induction chamber.
- Transfer the mice to the induction chamber and slightly anesthetize them.
- Inject subcutaneously 50 μl of parasites at 20 x 106/ml in one, or both, hind footpads per mouse using U-100 insulin syringe (final Lgy concentration: 106 parasites per footpad). Video 1. Luciferin injection and image acquisition demonstration. The video demonstrates how to use the isoflurane anesthesia system, injection of luciferin in mice (Step C2), mice anesthesia and transfer to SPFC chamber (Step C3) and image acquisition using the In-vivo Xtreme II system (Step C4). (All animal protocols in this publication were approved by the Swiss Federal Veterinary Office (SFVO), under the authorization number 2113. Animal handling and experimental procedures were undertaken with strict adherence to ethical guidelines set out by the SFVO and under inspection by the Department of Security and Environment of the State of Vaud, Switzerland.)
- Parasite burden quantification by Bioluminescence using In-Vivo Xtreme II
Parasite burden can be determined either immediately after parasite infection or along the time-course of the experiment. 5 mice at a time can be imaged.- Start the In-Vivo Xtreme II system and set up the Bioluminescent protocol.
- Switch on the apparatus.
- Wait for 2 min before starting the computer.
- Once the computer is on, access the MI software and select Capture Xtreme BI 4MP.
- Allow the camera to reach its working temperature of -90 °C. The CCD Operating Temperature box will change from red to green when the camera is ready to use.
Note: The duration of this step is approximate 10 min. - Input predetermined capture parameters (Table 1) (see Note):
Table 1. Capture values for the different modalities. Modality: imaging modes available in the In-Vivo Xtreme II (fluorescence, luminescence, X-Ray and reflectance). Exposure time: duration of time the camera collects light (seconds, minutes). Mode: way the camera captures the image. kVp: maximum tension applied across an X-Ray tube. Binning: process of combination of neighboring pixels on an image sensor into one “super pixel”. fSTOP: aperture of the diaphragm of the camera. FOV: Field of view. Focal plane: focus.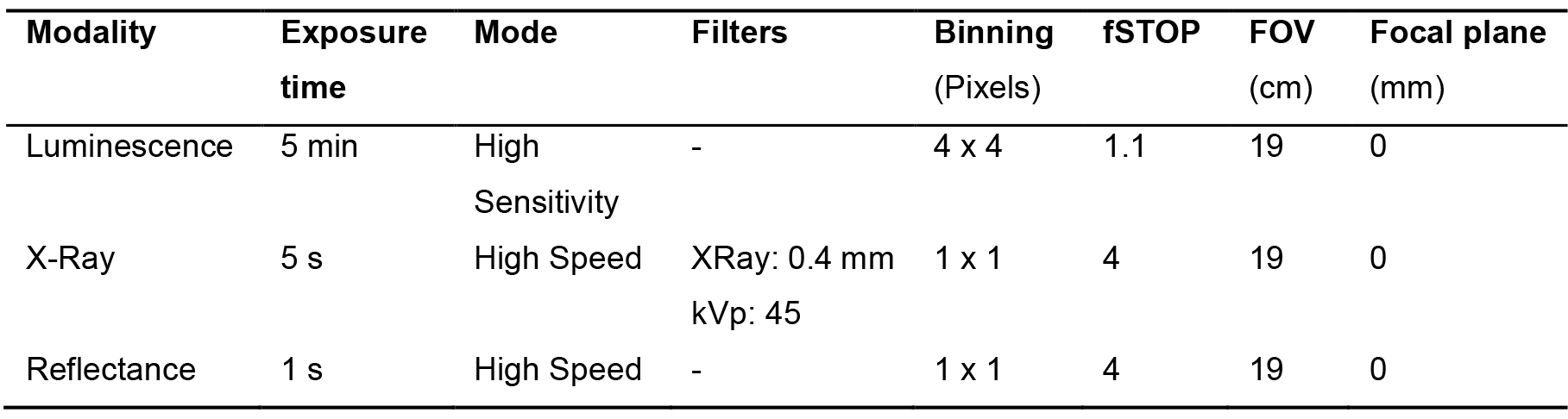
- Save the parameters in Step C1e as individual session files.
- Create a protocol by selecting the Create/Edit Protocols button from the system interface acquire window.
- Select the previously luminescence session file as Step C1. Next, select the corresponding X-Ray session file as Step C2. Finally, choose the reflectance session file as in Step C3. Keep–Do nothing–in both Before and After Image Capturer in the 3 Steps of the protocol.
- Save the protocol.
- Injection of luciferin solution in mice (Video 1, part 2)
- Inject intraperitoneally 200 μl of luciferin solution (Recipe 6) at a final dose of 150 mg/kg/ mouse using U-100 insulin syringe (see Note).
- Wait for 10 min after luciferin administration before starting image acquisition.
Note: Mice are not anesthetized during luciferin injection. Light of the laminar hood is turned off during injection of luciferin in mice.
- Isoflurane anesthesia and transfer to the SPFC chamber (Video 1, parts 3, 4, 5 and 6) 6-7 min after injection of luciferin in mice.
- Repeat Procedure B for mice anesthesia using the induction chamber.
- Transfer the anesthetized mice to the individual nosecones of the SPFC chamber connected to the isoflurane system (Figure 2A).
- Introduction of SPFC chamber into the In-Vivo Xtreme system and image acquisition (Video 1, parts 6, 7 and 8) 10 min after injection of luciferin in mice.
- Switch on the isoflurane system connected to the imaging system.
- Insert the SPFC chamber to the In-Vivo Xtreme machine.
- Connect all the isoflurane pipes to the SPFC (Figure 2B).
- Execute the previously saved protocol (Step C1i) by clicking Execute Protocol button from the main acquire window interface. During the execution of the protocol, notice a status window providing real time report of the current capture, the exposure duration as well as the protocol step.
- After image capture, remove the SPFC chamber from the imaging system and switch off the isoflurane system.
- Transfer the anesthetized mice to their cage and monitor the animals until they are fully recovered.
- Open the acquired images in the MI software for visual inspection and region of interest (ROI) analysis.
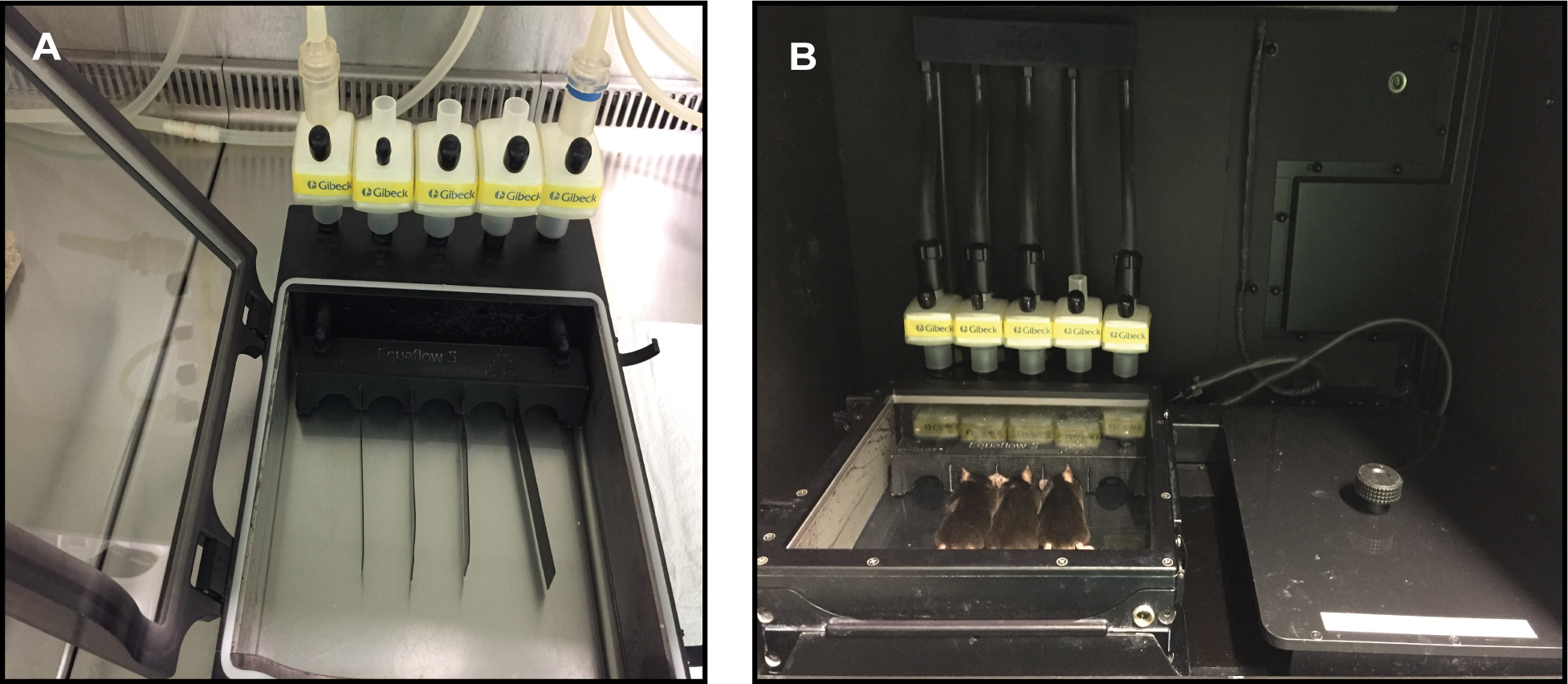
Figure 2. SPFC chamber. A. SPFC chamber connected to the isoflurane system for introducing the anesthetized mice. B. SPFC chamber at the Xtreme II system connected to the isoflurane system.
- Start the In-Vivo Xtreme II system and set up the Bioluminescent protocol.
Data analysis
- Image Overlay
Luminescence image overlay can be performed either with X-Ray or Reflectance image.
Example using Luminescence and X-Ray images:- Open the Luminescence (foreground) image with the MI software (Figure 3A).
- Remove the background of the Luminescence image by clicking on Image Display tab on the top bar and set the Min/Max values (Figure 3B).
Note: Min/Max range differs from study to study depending on the luminescence intensity captured. Images should be contrasted with a Min/Max that shows specific signal avoiding any unspecific background signal. - Convert to Photons/s/mm2 (Figure 3C).
- Open the X-Ray (background) image with the MI software (Figure 3D).
- Click the Overlay option from the Image Display tab, select the Luminescence image and choose the Transparency option (Figure 3E).
- To extract the overlayed image, select the Annotations option from the Navigation menu (Figure 3F). Add any additional data or object to the overlay. Once all the annotations are included, select the Select all option from the Edit top tab. Copy and paste the overlayed image into the desired file (e.g., Word, PowerPoint, Photoshop) for display.
- Image Analysis
Luminescence intensity quantification. 5 mice can be analyzed at a time.- Repeat steps1-4 from the Image Overlay section.
- Open the Manual ROIs tab from the Navigation panel and click on New ROI set (Figure 3F).
- Create an ROI set and adjust it to better enclose the luminescence intensity detected (Figure 3G).
Note: The ROI set can be saved as Template for the following image analysis using the same experiment layout. - To view the quantitative results, select the Analysis tab from the top Menu bar and select the Display tab (Figure 3H).
- From the Display option, select the desired parameters and then click OK.
- Copy the results obtained and paste them into the appropriate file for further analysis (e.g., Word, Excel, GraphPad Prism). e.g., Copy the Sum values from the Analysis tab (Figure 3H) to GraphPad Prism software. Represent the data as mean and ±SD or SEM (Figure 3I).
- Save the image so the ROI analysis is available.
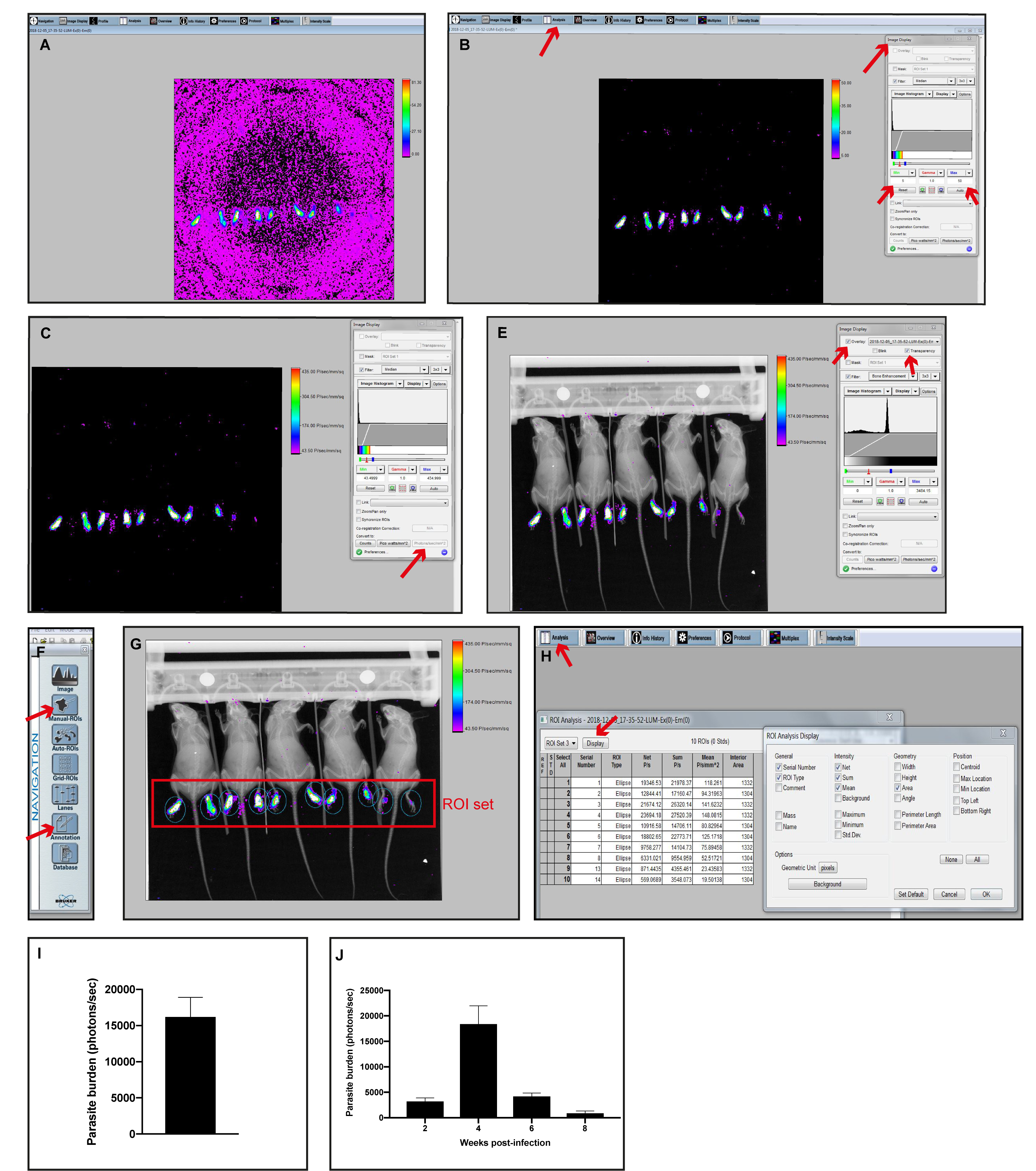
Figure 3. Monitoring the level of bioluminescence activity in the footpad after subcutaneous inoculation of 106 luciferase-transfected Lgy: an example of Image Overlay and Image Analysis. A-C. Step by step analysis of removing the background from a luminescence capture and converting the intensity to photons/sec/mm2. D. Example of X-Ray capture. E. Overlay of C + D images. F. Navigation display. G. Overlay with the set of ROIs. H. Quantitative results obtained from the Analysis tab. I. Parasite burden from Lgy infected mice footpads at 4 weeks post-infection (H Sum column, photons/sec) using GraphPad Prism software. J. Parasite burden kinetics from Lgy infected mice footpads (Sum, photons/sec). I-J. Data represent mean and ±SEM from 5 mice.
Notes
- Capture parameters were predetermined with the help of Bruker Corporation (BRUKER).
Luminescence modality: Luminescence signal is low in our experimental model. We use an exposure time of 5 min to increase the signal detection, but also taking into account to minimize the mice anesthesia. High sensitivity mode in order to reduce the background and increase the sensitivity of the luminescent signal. Binning of 4x4 increases the intensity of the signal but does not matter if it lowers the resolution of the image. Large aperture (fSTOP 1.1) helps to improve the detection of weak signal such as luminescence. FOV 19 allows imaging 5 mice/capture.
XRay and Reflectance modalities: Exposure time is in seconds and high-speed mode since the signal is strong. Binning 1x1 (no binning) and fSTOP 4 increase the resolution and the depth of field in the image, respectively. 45 kVp is used for X-Ray capture as it allows better bone resolution in vivo.
Recipes
- Complete parasite culture medium
500 ml Schneider’s Drosophila Medium
100 ml heat-inactivated FBS (Recipe 2)
5 ml HEPES
5 ml P/S
250 μl Biopterin (Recipe 3)
1 ml Hemin-folate (Recipe 4)
Filter all the reagents using Vacuum Filtration 500-Filtermax, aliquot and store at 4 °C - Heat-inactivated FBS
Thaw the FBS at RT. Then, perform heat-inactivation by incubating the FBS in a water bath at 56 °C for 30 min. Aliquot and store at -20 °C - Biopterin
Biopterin is reconstituted to a concentration of 1.2 mg/ml in DMSO. Aliquot and store at -20 °C - Hemin-folate
Dissolve 100 g hemin and 200 g folic acid in 20 ml 0.1 M NaOH
Add 40 ml of H2O
Adjust the pH to 8
Autoclave
Aliquot and store at 4 °C - Parasite fixative solution
48 ml DPBS
2 ml Glutaraldehyde solution technical
Store at 4 °C - Luciferin solution
66.6 ml DPBS
1 g VivoGloTM Luciferin
Luciferin is reconstituted to a concentration of 15 mg/ml in DPBS. Filter the solution using a 0.22 μm pore size filter and 50 ml syringe. Aliquot and store at -20 °C. Aliquots are thawed at RT prior to injection
Acknowledgments
We thank Slavica Masina for critical reading of the manuscript and Tiia Snäkä for the help with the video preparation. This research was supported by FNS grant No. 310030_173180 and No. IZRJZ3_164176/1 to NF and by aIRR to NF. This protocol was adapted from the protocol described in Ives et al. (2011) and Hartley et al. (2018).
Competing interests
The authors declare no competing interests.
Ethics
All animal protocols in this publication were approved by the Swiss Federal Veterinary Office (SFVO), under the authorization number 2113. Animal handling and experimental procedures were undertaken with strict adherence to ethical guidelines set out by the SFVO and under inspection by the Department of Security and Environment of the State of Vaud, Switzerland.
References
- Eren, R. O., Reverte, M., Rossi, M., Hartley, M. A., Castiglioni, P., Prevel, F., Martin, R., Desponds, C., Lye, L. F., Drexler, S. K., Reith, W., Beverley, S. M., Ronet, C. and Fasel, N. (2016). Mammalian innate immune response to a Leishmania-resident RNA virus increases macrophage survival to promote parasite persistence. Cell Host Microbe 20(3): 318-328.
- Hartley, M. A., Bourreau, E., Rossi, M., Castiglioni, P., Eren, R. O., Prevel, F., Couppié, P., Hickerson, S. M., Launois, P., Beverley, S. M., Ronet, C. and Fasel, N. (2016). Leishmaniavirus-dependent metastatic leishmaniasis is prevented by blocking IL-17A. PLoS Pathog 12(9): e1005852.
- Hartley, M. A., Eren, R. O., Rossi, M., Prevel, F., Castiglioni, P., Isorce, N., Desponds, C., Lye, L. F., Beverley, S. M., Drexler, S. K. and Fasel, N. (2018). Leishmania guyanensis parasites block the activation of the inflammasome by inhibiting maturation of IL-1β.Microb Cell 5(3): 137-149.
- Ives, A., Ronet, C., Prevel, F., Ruzzante, G., Fuertes-Marraco, S., Schutz, F., Zangger, H., Revaz-Breton, M., Lye, L. F., Hickerson, S. M., Beverley, S. M., Acha-Orbea, H., Launois, P., Fasel, N. and Masina, S. (2011). Leishmania RNA virus controls the severity of mucocutaneous leishmaniasis. Science 331(6018): 775-778.
- Lye, L. F., Owens, K., Shi, H., Murta, S. M., Vieira, A. C., Turco, S. J., Tschudi, C., Ullu, E. and Beverley, S. M. (2010). Retention and loss of RNA interference pathways in trypanosomatid protozoans. PLoS Pathog 6(10): e1001161.
- Rossi, M., Castiglioni, P., Hartley, M. A., Eren, R. O., Prével, F., Desponds, C., Utzschneider, D. T., Zehn, D., Cusi, M. G., Kuhlmann, F. M., Beverley, S. M., Ronet, C. and Fasel, N. (2017). Type I interferons induced by endogenous or exogenous viral infections promote metastasis and relapse of leishmaniasis. Proc Natl Acad Sci U S A 114(19): 4987-4992.
- van Griensven, J., Carrillo, E., Lopez-Velez, R., Lynen, L. and Moreno, J. (2014). Leishmaniasis in immunosuppressed individuals. Clin Microbiol Infect 20(4): 286-299.
Article Information
Copyright
© 2019 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Reverte, M. and Fasel, N. (2019). Leishmania Parasite Quantification by Bioluminescence in Murine Models. Bio-protocol 9(22): e3431. DOI: 10.21769/BioProtoc.3431.
Category
Microbiology > Pathogen detection > Bioluminescence
Cell Biology > Cell imaging > Confocal microscopy
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link