- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Assessing Different Ways of Bacillus subtilis Spreading over Abiotic Surfaces
Published: Vol 9, Iss 22, Nov 20, 2019 DOI: 10.21769/BioProtoc.3425 Views: 5513
Reviewed by: Juan Facundo Rodriguez AyalaChao JiangAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Purification of Native Dentilisin Complex from Treponema denticola by Preparative Continuous Polyacrylamide Gel Electrophoresis and Functional Analysis by Gelatin Zymography
Pachiyappan Kamarajan [...] Yvonne L. Kapila
Apr 5, 2024 2112 Views

Multiplexed Microfluidic Platform for Parallel Bacterial Chemotaxis Assays
Michael R. Stehnach [...] Jeffrey S. Guasto
Sep 5, 2024 2075 Views

In Silico Prediction and In Vitro Validation of Bacterial Interactions in the Plant Rhizosphere Using a Synthetic Bacterial Community
Arijit Mukherjee [...] Sanjay Swarup
Nov 5, 2025 1697 Views
Abstract
Surface-associate motility on biotic and abiotic environments is a key mechanism used by the model bacterium Bacillus subtilis and its closest relatives (i.e., B. amyloliquefaciens, B. thuringiensis, B. cereus, B. pumilus) for surface colonization and spreading across surfaces. The study of this mechanism in a research, industrial or clinic laboratory is essential; however, precautions should be taken for the reproducibility of the results, for example, the procedure to inoculate the bacteria on the testing plate, the humidity of the plate and the agar concentration. In this protocol, we describe, using Bacillus subtilis, how to perform these assays and, in addition, we show how by varying the agar concentration in the plate, you can make a first approximation of what type of motility has other bacterial species.
Keywords: BacillusBackground
Bacteria were considered for many years as unicellular organisms that function and disperse individually. However, in recent years, a growing number of studies show that these microorganisms live in societies where they interact with each other or with higher organisms (Palková, 2004; Lombardia et al., 2006; Lyons and Kolter, 2015; Clinton and Rumbaugh, 2016). These bacterial communities use different types of movements to migrate, adhere and spread over different surfaces, both biotic and abiotic (Kearns, 2010). This movement is related to the ability of bacteria to colonize a host in a beneficial or harmful way (pathogen) (Chaban et al., 2015; Gao et al., 2016). For example, these surface-associated movements allow the microorganism to move to the area of the root and leaf of a plant, where it can colonize and form biofilms, and thus avoid colonization by a phytopathogen (Hinsinger et al., 2008; Vacheron et al., 2013; Kan et al., 2017). Bacteria move through various mechanisms across moist surfaces. Depending on the circumstances, the model spore-forming bacterium B. subtilis can colonize surfaces using flagellum-driven single cell based movement (swimming), flagellum-dependent multicellular spreading (swarming), or growth-powered passive surface translocation flagellum-independent (sliding) (Kearns, 2010; Grau et al., 2015). The flagellar synthesis for swarming and swimming motility is carried out by the hag gene. On the other hand, The sliding type movement is dependent on the synthesis of exopolysaccharides (through the EPS operon) and the master sporulation regulator Spo0A (through the spo0A gene) (Kearns, 2010; Grau et al., 2015).
Motility assays are widely used in applied and basic studies, both for human health and for agriculture. However, the detailed methods are generally not precise and difficult to reproduce. In this protocol, we show how to test the different types of spreading using B. subtilis as a model, which can be adapted to other bacterial genera. All the precautions that should be taken when carrying out this type of experiments are also described in detail.
Materials and Reagents
- Pipette tips 2-200 µl Eppendorf® epT.I.P.S. (Eppendorf, catalog number: 022492039)
- Pipette tips 50-1,000 µl Eppendorf® epT.I.P.S. (Eppendorf, catalog number: 022492055)
- Petri dishes 100 x 15 mm 500/cs (Fisher Scientific, catalog number: FB0875713)
- Sterile 150 x 20 mm-culture tube (Fisher Scientific, catalog number: 14-961-33)
- Eppendorf® Safe-Lock 1.5 ml microcentrifuge tubes (Eppendorf, catalog number: 022363204) Cryovial (Simport, catalog number: T310-2A)
- Inoculating Loop (Sigma-Aldrich, catalog number: I0889)
- Drigalski spatula (3bscientific, model: 1010258)
- B. subtilis strains
- B. subtilis strains NCIB3610 and JH642 (Bacillus Genetic Stock Center, catalog numbers: 3A1 and 1A96)
The NCIB3610 strain is a wild-type undomesticated reference strain (derived from the Marburg strain) proficient in swimming, swarming and sliding motilities (Grau et al., 2015).
The JH642 is a domesticated laboratory strain derived from the strain 168, that in turns it is derived from NCIB3610, which is only proficient in swimming motility (Grau et al., 2015). - B. subtilis strains RG4365 (laboratory stock, Grau et al., 2015) and isogenic derivates.
The RG4365 strain is a wild-type undomesticated reference strain derived from the natto strain, proficient in sliding motility only (Grau et al., 2015).
- B. subtilis strains NCIB3610 and JH642 (Bacillus Genetic Stock Center, catalog numbers: 3A1 and 1A96)
- Luria Bertani (LB) broth (Sigma-Aldrich, catalog number: L3522)
- Magnesium sulfate heptahydrate (MgSO4·7H2O) (Sigma-Aldrich, catalog number: M1880)
- Potassium chloride (KCl) (Sigma-Aldrich, catalog number: P9541)
- Nutrient Broth (Difco, catalog number: BD234000)
- Agar (Sigma-Aldrich, catalog number: A1296)
- Glycerol (Sigma-Aldrich, catalog number: G5516)
- LB 1.5% (see Recipes 1)
- LB 0.3% agar (see Recipes 2)
- LB 0.7% agar (see Recipes 3)
- 50% glycerol (see Recipes 4)
- Difco Sporulation Medium (DSM) broth (see Recipes 5)
Equipment
- Erlenmeyer flask (Fisher Scientific, catalog number: FB5006000)
- Pipettor (Gilson, catalog number: F167300)
- Orbital Shaker (Thermo Fisher Scientific, model: MaxQ SHKE4000)
- Refrigerated incubator (Thermo Fisher Scientific, Thermo ScientificTM, model: HerathermTM General Protocol Microbiological Incubators, catalog number: 51028064)
- Freezers (-20 °C; So-Low Environmental Equipment) (Siemens, model: So-Low Ultra C85-22)
- Autoclave (Tuttnauer, model: 6690)
- Spectrophotometer (Paralwall, model: PWL3100)
- Stereomicroscope (Carl Zeiss Stemi, model: Stemi 2000)
- Fiber Optic Light Source (Carl Zeiss Stemi, model: KL1500LCD)
- Digital Camera (Canom, model: PowerShot A80).
- Bunsen burner (Humbolt, catalog number: H-5870)
- Vortex (Sigma-Aldrich, catalog number: Z755621)
Procedure
- High-yield spore production of Bacillus subtilis
- Maintaining axenic conditions of working, put a loop (10 µl) from the glycerol stock culture of the corresponding B. subtilis strain on an LB agar plate, streak the inoculum and incubate ON at 37 °C.
Note: Bacteria plated on LB agar plates can be stored at 4 °C for two or three weeks. However, for long-term storage, they should be cryopreserved in cultures supplemented with 25% glycerol. The addition of glycerol stabilizes frozen bacteria, preventing damage of cell membranes and keeping cells alive. The cryopreserved Bacillus can be stored stably at -80 °C for several years. To do this, grow B. subtilis for 48 h at 37 °C in DSM broth with shaking (150 rpm) to obtain a high titer of spores per ml. Perform a thermal shock at 80 °C for 15 min to the grown culture (heat-treatment kills all vegetative bacteria that did not sporulate). This heat-treated culture is mixed in equal parts with a 50% glycerol solution (“see Recipe 4”) and mixed gently. Freeze the tubes at -80 °C until use. In this way, the low temperature (-80 °C) and the intrinsic spore robustness will maintain the cryopreserved strain alive and viable for a long time period without affection of bacterial survival due to subsequent freeze and thaw cycles that you could perform in the future. To recover bacteria from your glycerol stock, open the tube and use a sterile loop or pipette tip to scrape some of the frozen bacteria off of the top. It is not needed to let the glycerol bacterial stock completely thaw. Always keep your glycerol bacterial stock on ice while the procedure is carried out. The biggest advantage of working with spores instead of vegetative is, apart from the greater stability over time of the spore, that the thermal treatment that is carried out (80 °C for 15 min) allows the elimination of possible microbial contaminants (that do not form spores) that could have grown or been introduced during handling bacterial cultures. - Put 2 ml of sterile DSM broth in a sterile 150 x 20 mm-tube. Inoculate medium with a single colony grown as indicated in the previous step. When it is necessary, add the corresponding antibiotic to the medium.
- Incubate for 48 h at 37 °C with shaking (150 rpm).
- To calculate the bacterial cellular yield (number cells, or colony forming units (CFU), per ml), make appropriate serial dilutions of the grown culture, and then plate them on LB agar plates and incubated for 24 h at 37 °C before CFU counting (see Procedure B).
- In parallel, check the purity of the grown culture by plating a loop on LB 1.5 % agar plates and incubated for 24 h at 37 °C.
- Maintaining axenic conditions of working, put a loop (10 µl) from the glycerol stock culture of the corresponding B. subtilis strain on an LB agar plate, streak the inoculum and incubate ON at 37 °C.
- Preparation of the serial dilutions
- To begin the procedure, take 100 µl of the cell suspension and add 900 µl of deionized sterile water. Shake, or vortex, the suspension well, and label as ‘10-1’.
Note: This step can be used to count both vegetative strains and spores. - Before the sample settles, take 100 µl of the cell suspension under axenic conditions with a pipette and transfer it to 900 µl deionized sterile water. Vortex the sample, and label as ‘10-2’.
- Repeat this dilution step, using 100 µl of the previous suspension and a 900 µl-deionized sterile water. Label these dilution tubes sequentially until reaching the appropriate dilution (e.g., tubes ‘10-7’).
- Spread out using a Drigalski spatula 100 µl of the dilutions 10-5 to 10-7 in LB agar plates and incubated for 24 h at 37 °C by triplicate.
- The number of CFU per milliliter is calculated with the following equation:
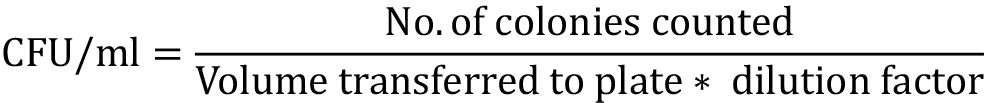
where,
No. of colonies counted = is the average of the number of colonies present in the three reading plates;
Volume transferred to the agar plates: is volume plate in ml (in this case it is 0.1 ml);
Dilution factor: is equal to the final volume divided by the aliquot volume.
- To begin the procedure, take 100 µl of the cell suspension and add 900 µl of deionized sterile water. Shake, or vortex, the suspension well, and label as ‘10-1’.
- Preparation of B. subtilis culture for spreading experiments
- Add 100 ml of non-sterile LB broth in an Erlenmeyer flask. Autoclave at 121 °C for 20 min. Cool flask and inoculate medium with 8 x 105 C.F.U./ml of a B. subtilis strain grown as indicated in Procedure A. Add the corresponding antibiotic to the medium in case of using isogenic mutant carrying antibiotic resistance.
Note: For the swarming and sliding experiments, the NCIB3610 strain was utilized instead of the JH642 strain, because the latter strain does not produce the biosurfactant surfactin (Grau et al., 2015) and therefore, it is unable to swarm or slide. - Incubate at mid-log at 37 °C (O.D. 575 = 0.5) with shaking (175 rpm). The number of C.F.U. per ml, for the strain JH642, at this growth phase will be around 5 x 107 to 7 x 107. If you use other bacterial strain, you must previously make a calibration (O.D. vs C.F.U.) growth curve to determine the precise O.D. that corresponds to the indicated C.F.U.
- Add 100 ml of non-sterile LB broth in an Erlenmeyer flask. Autoclave at 121 °C for 20 min. Cool flask and inoculate medium with 8 x 105 C.F.U./ml of a B. subtilis strain grown as indicated in Procedure A. Add the corresponding antibiotic to the medium in case of using isogenic mutant carrying antibiotic resistance.
- Preparation of plate for spreading experiments
- To assess the type of surface translocation (swarming, sliding) and/or swimming motilities, prepare LB fortified with 0.7% or 0.3% agar, respectively (Recipes 2 and 3).
- Cool the media at room temperature until ~50 °C approximately.
- Add 30 ml of the medium to a 10 cm-Petri dish (Figure 1, A-left part).
- Leave it to solidify at room temperature, not in the refrigerator.
- Dry for 1 h, in incubator at 37 °C (Figure 1, A-right part).
- When carrying out spreading assay, it is necessary to maintain the experimental conditions for the reproducibility of the experiments. The humidity of the media is one of the factors to control: very little water will result in low motility, while too much humidity can allow the swimming motility in undesired conditions. To control the moisture content, the plates are all prepared with agar medium at a relatively low temperature (~ 50 °C), and made of the same thickness. This prevents water loss by condensation in the lid of the plate. Finally, the spreading plates are dried all together, for the same time (~ 1 h), and in the same drying equipment (i.e., incubator at 37 °C). This procedure avoids, for example, that two plates have different degrees of humidity. The quality of the agar (i.e., manufacturer) affects the test; we recommend not to vary the type of agar during reproduction or repetition of the tests.
- Always use fresh plates, prepared not longer than 6 h before to perform the assay. Do not store the prepared plates.
- If the medium is supplemented, check that it does not affect the concentration of needed agar. If that is the case, add sterile water in the same proportion to the medium that has not been supplemented or prepare media with higher concentrations of agar that compensate this dilution.
- Spreading assay
- Using axenic procedures, inoculate the center of the Petri dish with a 1 µl drop containing, approximately, 6 x 107 cells/ml of the culture grown in Procedure C (Figure 1B).
Note: Deposit the drop on the agar, preventing the drop spattering or punch the agar with the tip. Other instruments can be used to inoculate such as wire inoculation needle or sterile toothpicks, however, they are not recommended because the volume inoculated is not precise and the agar can be easily damaged, which would affect the reproducibility of the technique. - Wait a few minutes for the drop to be absorbed in the agar before incubate (Figure 1C).
Note: The drop must be completely absorbed before incubation so that it does not spill on the agar. - Incubate at 37 °C for 48 h (Figure 1D).
Notes:- Do not stack the Petri dishes by more than two, so that the temperature transfer is the same in all the plates.
- The incubation temperature of the assays can be modified according to the experiment carried out and the bacteria used.
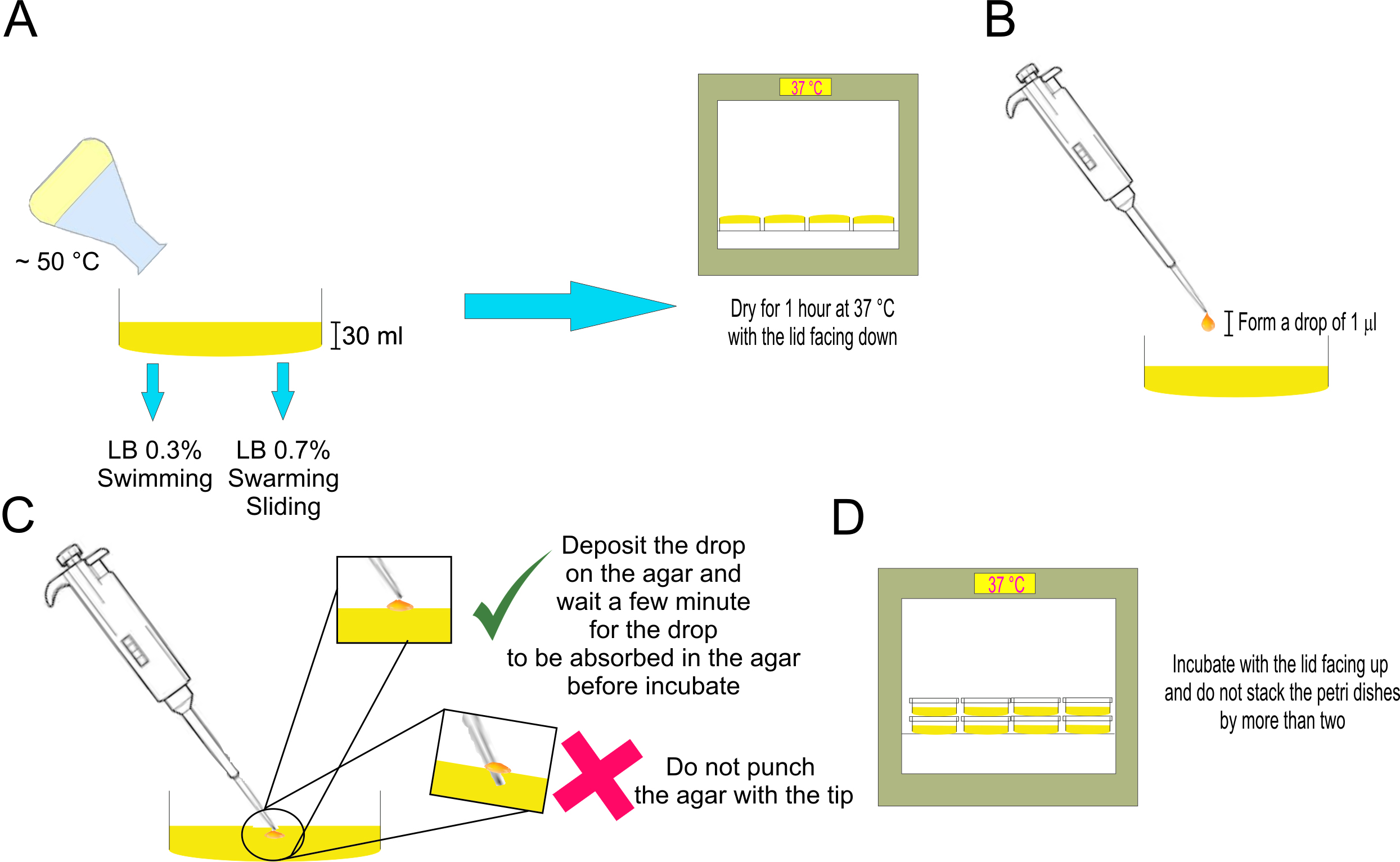
Figure 1. Diagram of the procedure to carry out a spreading assay. A. Scheme of preparation and drying of the plate to be used for the spreading experiments. B. Generation of the bacterial drop to inoculate the spreading plate. C. Correct and incorrect ways to deposit the drop on the culture medium of the spreading plate. D. Scheme of how to incubate the inoculated spreading plates ON. - The developed swimming, swarming, and sliding cellular discs are visualized using stereomicroscope and measuring the diameter of the colony (Figure 2A) as a function of time. Representative pictures from several independent experiments of top-viewed should be taken.
Note: Measure different colony diameters because the growths are usually not circular or uniform. Calculate the averages of the diameters of each time (for example X1 = (X1A + X1B)/2). - Graph the diameter of the bacterial spreading as a function of time (Figure 2B).
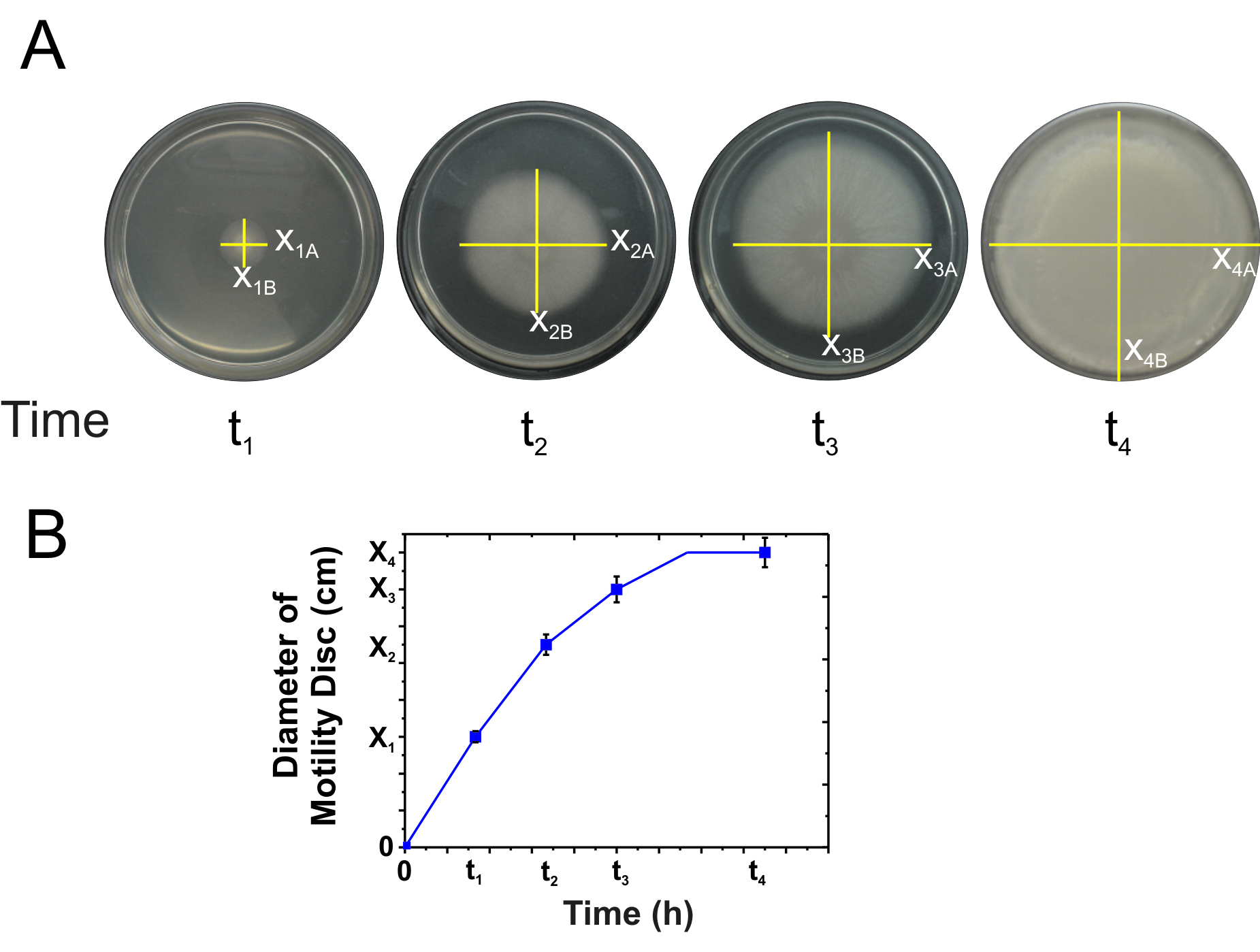
Figure 2. Diagram for the determination of the spreading as a function of time. A. Representative pictures from top-viewed of spreading bacteria over a testing plate.The yellow lines evidence the diameter of the colony. B. Graph of the average diameter (e.g., X1A + X1B/2 = X1) of the colony as a function of time. The values shown are mean ± SD of three independent times by quadruplicate.
Note: The appearance of the spreading of strains NCIB3610 and RG4365 is similar between them, being the spreading of NCIB3610 faster than the spreading of RG4365 (data not shown). The strain JH642 is not motile on surfaces over time (e.g., X1 = X2 = ..Xi..= Xn). Please refer to Grau et al. (2015) for the detailed information. - The bacterial strain is considered to be mobile when the diameter (d) of the colony is at least 20% greater than the diameter of the colony of the same strain developed in a medium that does not allow any movement over a solid surface (e.g., LB 1.5/2.0% agar) (Figure 3).
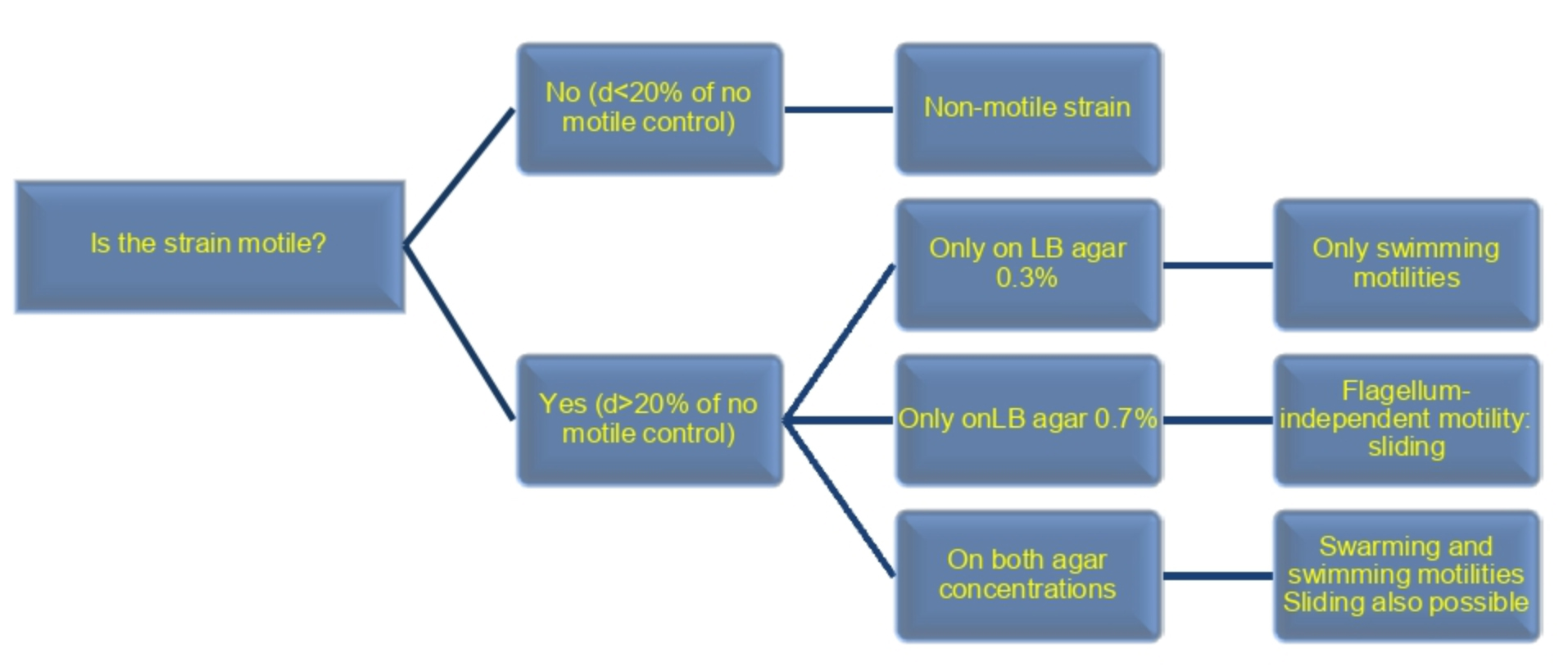
Figure 3. Analysis of results. Type of B. subtilis motility based on the results obtained. These results can be validated through genetic analysis, for example gene inactivation of hag genes (involved in flagellum synthesis) to confirm or discard flagella-mediated motility or inactivation of eps genes (involved in exopolysaccharide synthesis) to confirm or discard sliding motility.
Notes:- For Bacillus, these three types of motilities have been reported so far.
- Swarming (flagella-hag-dependent EPS-independent) and sliding (flagella-independent EPS-dependent) motilities can occur during development in the same bacterial colony. To know if these bacteria have one or both types of motilities, the following types of experiments can be performed. If you did not observe swimming (flagella-hag-dependent and EPS-independent) type motility but if it does occur in LB supplemented with 0.7% agar, you are facing a possible sliding (flagella-hag-independent and EPS-dependent) type motility. If you observed a proficient motility phenotype in LB supplemented with 0.3% and 0.7% agar concentrations, your strain may have one (sliding) or both types of motilities (sliding and swarming). In order to distinguish between swarming and sliding type motility, a mutation must be made that does not allow the synthesis of flagella (i.e., genetic inactivation of the hag gene), since swarming is flagellar motility dependent, while the sliding is not. Another way to distinguish between swarming and sliding, is the gene inactivation of sliding proficiency (i.e., gene inactivation of genes involved in EPS production that is essential for sliding, without affecting swarming proficiency).
Data analysis
- All experiments were performed using a randomized design.
- The values shown are mean ± SD of three independent times by quadruplicate.
- Use the Student’s t-test with a significance cut-off level of P < 0.05 for comparisons between two groups.
- Use the one-factor (ANOVA) variance analysis and correct by the post hoc Bonferroni test for multiple comparisons.
- Use the one-factor (ANOVA) to test the null hypothesis that there were no significant differences in the mean of s.d. of the seven strains, followed by Tukey´s post-hoc test.
- All statistical analyses were performed with GraphPad Prism version 5.0 (GraphPad Software, San Diego, CA).
Recipes
- Luria Bertani (LB) 1.5%
- Dissolve 20 g LB broth and 15 g agar to 1 L of dH2O
- Autoclave at 121 °C, 1 atm, for 20 min
- Store at room temperature
- Luria Bertani (LB) 0.3%
- Dissolve 20 g LB broth and 3 g agar to 1 L of dH2O
- Autoclave at 121 °C, 1 atm, for 20 min
- Store at room temperature
- Luria Bertani (LB) 0.7%
- Dissolve 20 g LB broth and 7 g agar to 1 L of dH2O
- Autoclave at 121 °C, 1 atm, for 20 min
- Store at room temperature
- 50% glycerol
- Dissolve 25 ml of pure glycerol (99.5 %) into 25 ml of dH2O
- Autoclave at 121 °C, 1 atm, for 20 min
- Store at room temperature
- DSM broth
- Dissolve 8 g nutrient broth, 1 g KCl and 0.25 g MgSO4 heptahydrate to 1 L of dH2O
- Autoclave at 121 °C, 1 atm, for 20 min
- Store at room temperature
Acknowledgments
The authors thank CONICET and FONCyT from Argentina, the Fulbright (Washington DC) and Pew (Philadelphia) foundations for their support and technical support. MB and RG are a postdoc fellow and carrier investigator from CONICET, respectively.
Competing interests
The authors have no conflict of interest to report.
References
- Chaban, B., Hughes, H. V. and Beeby, M. (2015). The flagellum in bacterial pathogens: For motility and a whole lot more. Semin Cell Dev Biol 46: 91-103.
- Clinton, A. and Rumbaugh, K.P. (2016) Interspecies and interkingdom signaling via quorum signals. Isr J Chem 56: 265–272.
- Gao, S., Wu, H., Yu, X., Qian, L. and Gao, X. (2016) Swarming motility plays the major role in migration during tomato root colonization by Bacillus subtilis SWR01. Biological Control 98: 11–17.
- Grau, R. R., de Ona, P., Kunert, M., Lenini, C., Gallegos-Monterrosa, R., Mhatre, E., Vileta, D., Donato, V., Holscher, T., Boland, W., Kuipers, O. P. and Kovacs, A. T. (2015). A duo of potassium-responsive histidine kinases govern the multicellular destiny of Bacillus subtilis. MBio 6(4): e00581.
- Hinsinger, P., Bravin, M.N., Devau, N., Gerard, F., Le Cadre, E., and Jaillard, B. (2008). Soil-Root-Microbe interactions in the rhizosphere: a key to understanding and predicting nutrient bioavailability to plants. R.C. Suelo Nutr. Veg 8: 39-47.
- Kan, J., Fang, R. and Jia, Y. (2017). Interkingdom signaling in plant-microbe interactions. Sci China Life Sci 60(8): 785-796.
- Kearns, D. B. (2010). A field guide to bacterial swarming motility. Nat Rev Microbiol 8(9): 634-644.
- Lombardia, E., Rovetto, A. J., Arabolaza, A. L. and Grau, R. R. (2006). A LuxS-dependent cell-to-cell language regulates social behavior and development in Bacillus subtilis. J Bacteriol 188(12): 4442-4452.
- Lyons, N. A. and Kolter, R. (2015). On the evolution of bacterial multicellularity. Curr Opin Microbiol 24: 21-28.
- Palková, Z. (2004). Multicellular microorganisms: laboratory versus nature. EMBO Rep 5(5): 470-476.
- Vacheron, J., Desbrosses, G., Bouffaud, M. L., Touraine, B., Moenne-Loccoz, Y., Muller, D., Legendre, L., Wisniewski-Dye, F. and Prigent-Combaret, C. (2013). Plant growth-promoting rhizobacteria and root system functioning. Front Plant Sci 4: 356.
Article Information
Copyright
© 2019 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Bartolini, M. and Grau, R. (2019). Assessing Different Ways of Bacillus subtilis Spreading over Abiotic Surfaces. Bio-protocol 9(22): e3425. DOI: 10.21769/BioProtoc.3425.
Category
Microbiology > Microbe-host interactions > Bacterium
Microbiology > Microbial cell biology > Cell motility
Cell Biology > Cell movement > Cell motility
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











