- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Using Imaging Flow Cytometry to Characterize Extracellular Vesicles Isolated from Cell Culture Media, Plasma or Urine
Published: Vol 9, Iss 21, Nov 5, 2019 DOI: 10.21769/BioProtoc.3420 Views: 4899
Reviewed by: Xiaoyi ZhengBegona DiazAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Isolation and Ex Vivo Testing of CD8+ T-Cell Division and Activation Using Mouse Splenocytes
Melissa Dolan [...] John M.L. Ebos
Aug 20, 2025 3864 Views

Detection of Autophagy in Human Peripheral Blood Mononuclear Cells Using Guava® Autophagy and Flow Cytometry
Melanie Scherer [...] Jörg Bergemann
Sep 20, 2025 1413 Views

Protocol for the Isolation and Analysis of Extracellular Vesicles From Peripheral Blood: Red Cell, Endothelial, and Platelet-Derived Extracellular Vesicles
Bhawani Yasassri Alvitigala [...] Lallindra Viranjan Gooneratne
Nov 5, 2025 1442 Views
Abstract
The ability to non-invasively detect specific damage to the kidney has been limited. Identification of extracellular vesicles released by cells, especially when under duress, might allow for monitoring and identification of specific cell types within the kidney that are stressed. We have adapted a previously published traditional flow cytometry method for use with an imaging flow cytometer (Amnis FlowSight) for identifying EV released by specific cell types and excreted into the urine or blood using markers characteristic of particular cells in the kidney. Here we present a protocol utilizing the Amnis FlowSight Imaging Flow Cytometer to identify and quantify EV from the urine of patients with essential hypertension and renovascular disease. Notably, EV isolated from cell culture media and plasma can also be analyzed similarly.
Keywords: ExosomesBackground
Extracellular vehicles (EVs) are released from cells under normal conditions and their numbers are known to increase when the cells are exposed to stress conditions. Therefore, levels of urinary EVs have been shown to be associated with various kidney disorders such as polycystic kidney disease (Hogan et al., 2009), acute kidney injury (Aghajani Nargesi et al., 2017; Cappuccilli et al., 2018), and various glomerular diseases (Zhang et al., 2019).
We have previously shown that levels of EVs that are increased in patients with hypertension likely originate from podocytes (Kwon et al., 2017) and peritubular capillaries (PTC) (Sun et al., 2018; Zhang et al., 2019). We have also recently shown that urinary levels of EVs reflecting renal cellular senescence are altered in these patients (Santelli et al., 2019). We describe here a more detailed protocol that can be used for the isolation and quantitative characterization of EVs isolated from cell culture media, plasma, and urine (Kwon et al., 2017; Sun et al., 2018; Conley et al., 2019; Zhang et al., 2019). The primary advantages of this method of EV isolation are shorter processing time and obviating the requirement for an ultracentrifuge to isolate the EVs.
Materials and Reagents
- Opaque Black 1.5 ml microcentrifuge tubes (Midwest Scientific, catalog number: T7100BK)
- Avant clear, 1.5 ml microcentrifuge tubes (Midwest Scientific, catalog number: AVSS1700)
- dPBS (Life Technologies, catalog number: 14190-250)
- Albumin, bovine serum, fraction V (Sigma, catalog number: A3059)
- InvitrogenTM Total Exosome Isolation reagents
- From Urine (Thermo Fisher Scientific, Inc., catalog number: 4484452)
- From Cell culture media (Thermo Fisher Scientific, Inc., catalog number: 4478359)
- From Plasma (Thermo Fisher Scientific, Inc., catalog number: 4484450)
- From Serum (Thermo Fisher Scientific, Inc., catalog number: 4478360)
- Tag-It VioletTM proliferation and cell tracking dye (Biolegend, Inc., catalog number: 425101)
- PV1/PL-VAP, FITC, anti-human antibody (Lifespan Biosciences, catalog number: LS-C209607-100)
- CD31, Brilliant Violet 605, Anti-Human antibody (BioLegend, Inc., catalog number: 303122)
- CD144, PE, anti-human antibody (BioLegend, Inc., catalog number: 348506)
- Bleach, Clorox (Solution used in the FlowSight during operation)
- Isopropanol, HPLC grade (Honeywell, catalog number: AH323-4) (Solution used in the FlowSight during operation)
- Coulter Clenz® (BD Biosciences, catalog number: 8546929) (Solution used in the FlowSight during operation)
- Deionized H2O (Solution used in the FlowSight during operation)
- Molecular ProbesTM AbC Total Antibody compensation bead kit (Thermo Fisher Scientific, Inc., catalog number: A10497)
- Sucrose (Sigma, BioXtra, catalog number: S7903)
- MES: 2-(N-Morpholino) ethanesulfonic acid, 4-Morpholineethanesulfonic acid (Sigma, catalog number: M3671)
- DMSO
- EV Buffer (pH 7.4) (see Recipes)
- 5 mM Tag-It VioletTM (TIV) solution (see Recipes)
Equipment
- 1 ml pipettor
- FlowSight Imaging flow cytometer (Amnis, Inc.) equipped with:
- 488 nm Laser
- 405 nm Laser
- 642 nm laser
- 785 nm laser (SSC)
- Quantitative imaging
- 96 well plate autosampler
- Refrigerated Microcentrifuge (Eppendorf, Model 5415R, 48 x 2 ml rotor)
- MultiTherm incubated vortexer (Midwest Scientific, catalog number: MTHC-1500)
- Barnstead deionized water system (Model)
- -80 °C freezer
- -20 °C freezer
- 4 °C refrigerator
Software
- Amnis® IDEAS version 6.2 (Luminex Inc., https://www.luminexcorp.com/imaging-flow-cytometry/), data analysis software
- Amnis® INSPIRE Version 6, data acquisition software
- Excel (Microsoft)
- JMP version 10.0 (SAS Institute)
Procedure
- EV isolation
Note: EVs were isolated from whole urine using Total Exosome Isolation reagent (Invitrogen, Waltham, MA) according to the manufacturer’s guidelines.- Urine samples (1,000 μl) were centrifuged at 2,000 x g for 30 min at 4 °C to remove cells and debris.
- Urine supernatants (800 μl) were mixed with 1 volume (800 μl) of the Total Exosome Isolation reagent and incubated for 1 h at room temperature.
Other samples (Plasma, serum, cell culture media) can be prepared similarly using the manufacturer’s recommendations for reagent volume, incubation time, and temperature. - After incubation, samples were centrifuged at 10,000 x g for 1 h at 4 °C.
- Supernatant was removed using 1 ml pipettor and discarded.
- Pelleted exosomes were resuspended in 50 μl EV buffer (2 mol/L sucrose and 500 mmol/L MES [2-(N-morpholino) ethanesulfonic acid, 4-morpholineethanesulfonic acid] solution).
- Store at -80 °C. Samples are considered to be stable for at least 6 months (Mendt et al., 2018).
- EV staining and antibody labeling
- Start up and Calibrate FlowSight.
- Thaw samples on ice.
- Prepare 5 mM solution of Tag-It VioletTM by adding 50 μl DMSO to one tube of Tag-It VioletTM as in the Recipe 2 Tag-It Violet marks the EV by binding to the interior proteins, thus labeling the EVs, which are too small for brightfield visualization.
- Pipet 20 μl EVs using 20 μl pipettor into 1.5 ml opaque Black microcentrifuge tubes, to protect samples from light. The incubated vortexer has a clear lid and is often located in the main laboratory, thus the black tubes protect the fluorescent dyes from photobleaching during the incubations.
Include one sample of EVs that will be stained only with Tag-It Violet for single color compensation control. - Add 30 μl dPBS to each sample using a 100 μl pipettor.
- Add 0.5 μl of 5 mM TIV stock to EVs (50 μM final concentration) using a 2 μl pipettor.
- Incubate EV for 2 h at 37 °C at 250 rpm using MultiTherm incubated vortexer.
- Add antibodies (for example peritubular capillary identification) using a 10 μl pipettor.
3 μl PL-VAP(LSBio)
4 μl CD31 (Biolegend)
3 μl CD144 (Biolegend)
Incubate for 1 h at RT at 250 rpm in MultiTherm incubated vortexer.
- Data acquisition using FlowSight
- Acquisition is performed using FlowSight Imaging Flow Cytometer (Amnis Corporation, Seattle, WA) equipped with Amnis INSPIRE` software.
- Vortex sample for 10-20 s before loading onto FlowSight.
- Instrument setup:
Lasers (all set to full power):
405 nm 100 mW
488 nm 60 mW
642 nm 100 mW
785 nm (SSC) 70 mW - Acquire at least 100,000 TIV positive events.
- Prepare single color control compensation beads for antibodies.
- In 1.5 ml microcentrifuge tubes add 1 drop AbC capture beads.
- Add one antibody per tube with volume indicated in Step B8.
- Incubate at Room Temp for 15 min protected from light.
- After adding 1 ml dPBS, vortex and spin at 250 x g for 5 min using a 1000 μl pipettor.
- Remove supernatant.
- Add one drop AbC negative beads and 50 μl dPBS.
- Vortex and run on FlowSight.
- Acquire 1000 events with Brightfield off (for compensation).
- Acquire 1000-5000 events with Brightfield on to aid in initial gate determination.
- Samples are run in the following order.
- TIV only EVs
- Acquire 1000 events with Brightfield off (for compensation).
- Acquire 1000-5000 events with Brightfield on to aid in initial gate determination.
- Antibody single color controls (acquired as in Step C5g above)
- Samples with TIV and all antibodies (Brightfield is on)
- TIV only EVs
Data analysis
- Amnis IDEAS data analysis software
- EVs were quantified using IDEAS (Amnis, Seattle, WA).
- Compensation matrix for experiment is created using IDEAS compensation matrix wizard.
- Raw data file from one sample is opened and compensation matrix applied.
- Gating strategy template is created using stepwise data analysis wizard in IDEAS software based on antibody and Tag-It Violet staining panel. Figure 1 illustrates the gating strategy steps taken in flow cytometry data analysis for PTC identification.
For PTC identification, Tag-It Violet+/PL-VAP+/CD31-/CD144- events were of primary interest and the gating strategy was developed with that goal. The combination of PL-VAP+/CD31-/CD144- is used for identification of EV released from peritubular capillaries.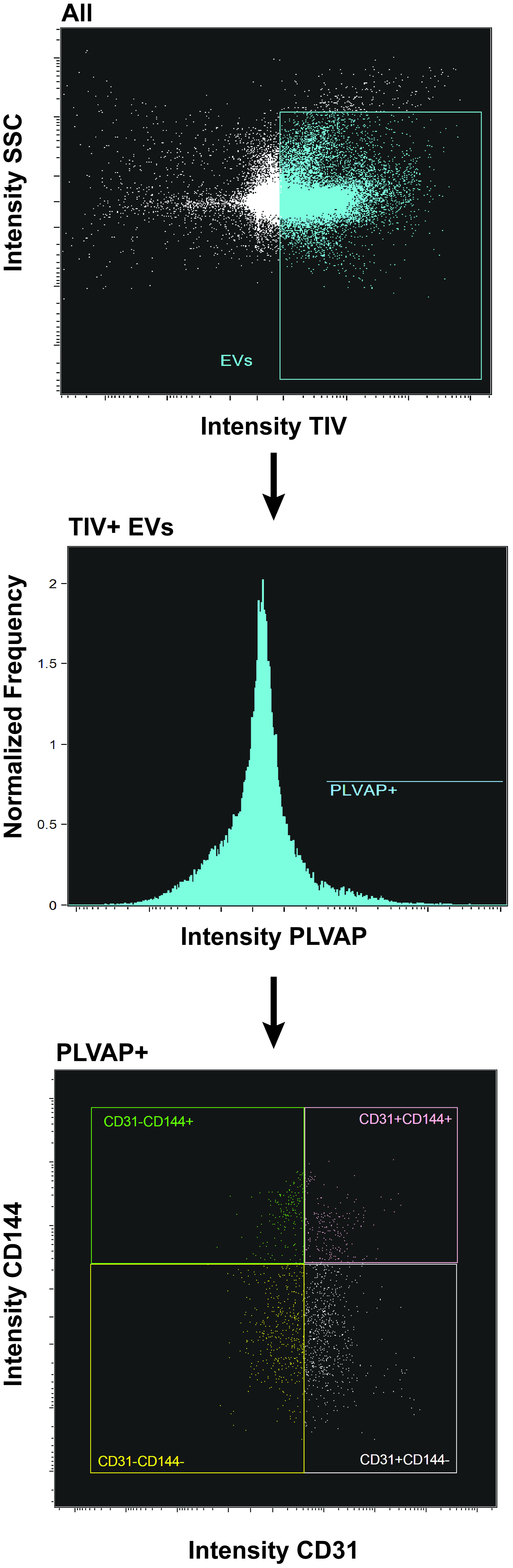
Figure 1. A schematic showing flow cytometry gating strategy for PTC identification. Step 1: Scatterplot of side scatter (size) and intensity of Tag-It Violet (EVs) from all events collected, Draw gate (ROI) for Tag-It violet positive events and SSC low; Step 2: Histogram of PL-VAP antibody intensity of the EV+ population, draw gate (ROI) for PL-VAP positive events; Step 3: Scatterplot of CD31 and CD144 from PL-VAP+ population, most important gate (ROI) being CD31-/CD144- for PTC EV, however gates (ROIs) for identification of CD31-/CD144+, CD31+, CD144-, and CD31+/CD144+ events are also drawn. - EVs are measured using the following flow-gating strategy (see Figure 1).
- Create Scatterplot of Side-scatter (SSC) channel 6 versus Tag-It Violet channel 7 of all events collected.
- Draw a region of interest (ROI) for positive Tag-it violet events and low SSC
- Label as Tag-It Violet+ or EV+ population
- Create Histogram of PL-VAP channel.
- Draw an ROI for positive PL-VAP
- Label as PL-VAP+
- Create Scatterplot of CD31 versus CD144
- Draw an ROI around events that are for negative CD31 and negative CD144
- Label as CD31-/CD144-
- Also draw ROIs for CD31+/CD144+ events
- Create Scatterplot of Side-scatter (SSC) channel 6 versus Tag-It Violet channel 7 of all events collected.
- Gating strategy with all ROIs is saved as a template within IDEAS.
- Saved template and compensation matrix is applied to all samples.
- Manual verification of ROIs on 3-5 samples was done to verify ROI border thresholds
- Data as number of events in each ROI from every sample was extracted.
- The gated populations are expressed as a percentage of Tag-It Violet positive events. (See Sun et al. (2018), Figures 2A and 2C, for comparison of data between groups of patients.)
- PL-VAP+/CD31-/CD144- events
- PL-VAP+/CD31+
- PL-VAP+/CD144+
- PL-VAP+/CD31+/CD144+
- Statistical analysis using JMP; please see Sun et al. (2018) for data analysis.
Recipes
- EV Buffer (pH 7.4)
2 mol/L sucrose (Sigma, BioXtra)
500 mmol/L MES (Sigma)
Stable for 6 months at 4 °C - 5 mM Tag-It VioletTM (TIV) solution
50 μl anhydrous DMSO
One vial of Tag-It VioletTM (122.25 μg)
Stable for 1 month at -20 °C, but best practice is to make fresh
Acknowledgments
This protocol was adapted from Sun et al. (2018). This work was partly supported by the National Institutes of Health grant numbers DK10081, DK104273, DK102325, and DK120292.
Competing interests
No competing interests.
Ethics
This study was approved by the institutional review board of the Mayo Clinic, and performed in accordance with the ethical principles of the Declaration of Helsinki. Informed written consent was obtained from each patient.
References
- Aghajani Nargesi, A., Lerman, L. O. and Eirin, A. (2017). Mesenchymal stem cell-derived extracellular vesicles for kidney repair: current status and looming challenges. Stem Cell Res Ther 8(1): 273.
- Cappuccilli, M., Capelli, I., Comai, G., Cianciolo, G. and La Manna, G. (2018). Neutrophil gelatinase-associated lipocalin as a biomarker of allograft function after renal transplantation: evaluation of the current status and future insights. Artif Organs 42(1): 8-14.
- Conley, S. M., Shook, J. E., Zhu, X. Y., Eirin, A., Jordan, K. L., Woollard, J. R., Isik, B., Hickson, L. J., Puranik, A. S., and Lerman, L. O. (2019). Metabolic syndrome induces release of smaller extracellular vesicles from porcine mesenchymal stem cells. Cell Transplant (unpublished).
- Hogan, M. C., Manganelli, L., Woollard, J. R., Masyuk, A. I., Masyuk, T. V., Tammachote, R., Huang, B. Q., Leontovich, A. A., Beito, T. G., Madden, B. J., Charlesworth, M. C., Torres, V. E., LaRusso, N. F., Harris, P. C. and Ward, C. J. (2009). Characterization of PKD protein-positive exosome-like vesicles. J Am Soc Nephrol 20(2): 278-288.
- Kwon, S. H., Woollard, J. R., Saad, A., Garovic, V. D., Zand, L., Jordan, K. L., Textor, S. C. and Lerman, L. O. (2017). Elevated urinary podocyte-derived extracellular microvesicles in renovascular hypertensive patients. Nephrol Dial Transplant 32(5): 800-807.
- Mendt, M., Kamerkar, S., Sugimoto, H., McAndrews, K. M., Wu, C. C., Gagea, M., Yang, S., Blanko, E. V. R., Peng, Q., Ma, X., Marszalek, J. R., Maitra, A., Yee, C., Rezvani, K., Shpall, E., LeBleu, V. S., and Kalluri, R. (2018) Generation and testing of clinical-grade exosomes for pancreatic cancer. JCI Insight 3(8):e99263.
- Santelli, A., Sun, I. O., Eirin, A., Abumoawad, A. M., Woollard, J. R., Lerman, A., Textor, Stephen C., Puranik, A. S. and Lerman, L. O. (2019). Senescent kidney cells in hypertensive patients release urinary extracellular vesicles. J Am Heart Assoc 8(11): e012584.
- Sun, I. O., Santelli, A., Abumoawad, A., Eirin, A., Ferguson, C. M., Woollard, J. R., Lerman, A., Textor, S. C., Puranik, A. S. and Lerman, L. O. (2018). Loss of renal peritubular capillaries in hypertensive patients is detectable by urinary endothelial microparticle levels. Hypertension 72(5): 1180-1188.
- Zhang, L. H., Zhu, X. Y., Eirin, A., Nargesi, A. A., Woollard, J. R., Santelli, A., Sun, I. O., Textor, S. C. and Lerman, L. O. (2019). Early podocyte injury and elevated levels of urinary podocyte-derived extracellular vesicles in swine with metabolic syndrome: role of podocyte mitochondria. Am J Physiol Renal Physiol 317(7): F12-F22.
Article Information
Copyright
© 2019 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Woollard, J. R., Puranik, A., Jordan, K. L. and Lerman, L. O. (2019). Using Imaging Flow Cytometry to Characterize Extracellular Vesicles Isolated from Cell Culture Media, Plasma or Urine. Bio-protocol 9(21): e3420. DOI: 10.21769/BioProtoc.3420.
Category
Molecular Biology > Protein > Flow Cytometry
Cell Biology > Cell-based analysis > Flow cytometry
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link