- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Time-lapse Imaging of Alveologenesis in Mouse Precision-cut Lung Slices
Published: Vol 9, Iss 20, Oct 20, 2019 DOI: 10.21769/BioProtoc.3403 Views: 7980
Reviewed by: Zinan ZhouThirupugal GovindarajanKarthik KrishnamurthyAnita Umesh

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Estrous Cycle Monitoring in Mice with Rapid Data Visualization and Analysis
Leanna K. Pantier [...] Catherine A. Christian
Sep 5, 2019 9699 Views
Abstract
Alveoli are the gas-exchange units of lung. The process of alveolar development, alveologenesis, is regulated by a complex network of signaling pathways that act on various cell types including alveolar type I and II epithelial cells, fibroblasts and the vascular endothelium. Dysregulated alveologenesis results in bronchopulmonary dysplasia in neonates and in adults, disrupted alveolar regeneration is associated with chronic lung diseases including COPD and pulmonary fibrosis. Therefore, visualizing alveologenesis is critical to understand lung homeostasis and for the development of effective therapies for incurable lung diseases. We have developed a technique to visualize alveologenesis in real-time using a combination of widefield microscopy and image deconvolution of precision-cut lung slices. Here, we describe this live imaging technique in step-by-step detail. This time-lapse imaging technique can be used to capture the dynamics of individual cells within tissue slices over a long time period (up to 16 h), with minimal loss of fluorescence or cell toxicity.
Keywords: Imaging alveologenesisBackground
Prenatal and postnatal lung development is classified into several distinct stages beginning with budding from the foregut endoderm followed by branching morphogenesis, sacculation and alveologenesis within surrounding lung mesenchyme. This developmental process is tightly regulated by a well-orchestrated signaling programme and cellular components (Kotton and Morrisey, 2014; Akram et al., 2016). The major function of the lungs is gas exchange, which occurs via diffusion (Herriges and Morrisey, 2014). This diffusion takes place between the thin cellular layers of alveolar epithelium and capillary endothelium (Roth-Kleiner and Post, 2005). Dysregulated alveologenesis is linked with a number of neonatal and infant diseases, including bronchopulmonary dysplasia (BPD) and pulmonary hypoplasia (Kreiger et al., 2006; Hilgendorff et al., 2014). In adults, alveolar damage is a component of several chronic lung diseases such as chronic obstructive pulmonary disease (COPD) and idiopathic pulmonary fibrosis (IPF). Currently, there are no curative treatments for these diseases other than lung transplantation (Warburton et al., 2006; Madurga et al., 2013; McGowan, 2014) and there is an unmet need to understand the mechanisms of alveologenesis in order to develop effective treatments.
In mice, sacculation begins around embryonic day 17.5 and is followed by alveologenesis, which begins within the first few days of postnatal life and is mostly completed in first month of life (Herriges and Morrisey, 2014). However, the most active phase of alveologenesis occurs in first two weeks of postnatal life with the majority of alveoli formed by postnatal day (P) 21 (Hind et al., 2002; Snoeck, 2015).
Current understanding, based on static imaging experiments, is that alveologenesis occurs through repeated septation events that sub-divide primary air sacs to increase the number and surface area of alveoli (Amy et al., 1977; Mund et al., 2008). Real-time visualization of alveologenesis is challenging due to their location deep inside the body and the relatively slow duration of this process. A recent study used both ex vivo and in vivo live-imaging to study the sacculation stage of mouse lung development, immediately prior to alveologenesis, but these techniques are not suitable for imaging postnatal lungs (Poobalasingam et al., 2017; Li et al., 2018).
Precision cut lung slices (PCLS) contain intact alveoli and are increasingly used to study lung biology and disease pathogenesis (Meng et al., 2008; Sanderson, 2011; Thornton et al., 2012). Time-lapse imaging of PCLS has been used to show dynamic interactions of mesenchymal cells and macrophages with the extracellular matrix in adult normal and fibrotic mouse lungs, as well as in PCLS of human lungs (Burgstaller et al., 2015). In addition, quantifiable ex vivo alveologenesis has been demonstrated in early postnatal mouse PCLS culture (Pieretti et al., 2014). Using a combination of widefield microscopy and image deconvolution on postnatal mouse PCLS we have developed a method to capture the morphological mechanisms of alveologenesis in real-time (Akram et al., 2019). Here we describe the detailed protocol for real-time live imaging of postnatal alveologenesis.
Materials and Reagents
- 50 ml centrifuge tubes (Thermo Fisher, catalog number: 338652)
- Metallic spatula (Fisher Scientific, catalog number: 11523482)
- Glass coverslips (Thermo Fisher, catalog number: 102260)
- Probe Point (Blunt) needles, 25 G, 19 mm (0.75 inch) (Harvard Apparatus, catalog number: 725461) for P3 mice; Monoject blunt needles with Aluminum Hub, 23 G, 1 inch (Harvard Apparatus, catalog number: 722349) for P7 mice, and 21 G (Custom made from 21 G syringe needle) for P14 and adult mice (all from Harvard Apparatus UK)
- 24-well plate (Corning® Costar® TC-Treated Multiple Well Plates) (Sigma-Aldrich, CLS3527-100EA)
- 96-well plate (Thermo Fisher, catalog number: 249952)
- Ibidi 24-well μ-plate (Uncoated) (Ibidi, catalog number: 82401)
- Transwell (0.4 µm pore, 12 mm, polyester membrane) (Corning, catalog number: 3460)
- Microscope slides, SuperFrost®, Menzel Gläser (VWR, catalog number: 631-1318)
- Non-sterile silk black braided suture spool, 22.9 m, Size 5-0 (Harvard Apparatus UK, catalog number: 517607)
- BD Micro-Fine+ 29 G, 1 ml Insulin Syringes (MediSuuplies, catalog number: PMC3743)
- Syringes, 5 ml (VWR International, catalog number: SART16644-E)
- Thermo ScientificTM NuncTM Cell Culture/Petri Dishes, 100 mm (Fisher Scientific, catalog number: 10508921)
- Swann-Morton Surgical scalpels, No.22 (MediSupplies, catalog number: PMC0105)
- Wet ice and ice box
- C57BL/6 male and female mice from Charles River Laboratories
- EpCAM-FITC (CD326) monoclonal antibody (eBioscience, catalog number: 11-5791-80; Clone G8.8)
- Alexa-647 conjugated PECAM antibody (CD31-Alexa 647) (Biolegend, catalog number: 102416; Clone 390)
- Laboratory tissue (Blue roll)
- Metal flat washer (weight 1.66 g) (M8-5/16th inches diameter) (B&Q, UK)
- Absolute ethanol (Sigma-Aldrich, catalog number: 34852-M)
- Pentobarbitone (Pentoject, Animalcare, catalog number: XVD 132)
- Low-melting-point agarose (Sigma-Aldrich, catalog number: A9414)
- Hanks' Balanced Salt Solution (HBSS) (1x) (Life Technologies, catalog number: 14025-050)
- HEPES 1 M (Life Technologies, Gibco, catalog number: 15630080)
- Phosphate Buffered Saline (PBS) (Life Technologies, Gibco, catalog number: 20012068)
- Dulbecco's Modified Eagle Medium (DMEM) (Life Technologies, Gibco, catalog number: 31966-021), also referred to as 'DMEM basal media” in the Recipes
- Penicillin-Streptomycin (10,000 U/ml) (Life Technologies, Gibco, catalog number: 15140122)
- MTT reagent (Thiazolyl Blue Tetrazolium Bromide) (Sigma-Aldrich, catalog number: M2128)
- Dimethyl sulfoxide (DMSO) (Sigma-Aldrich, catalog number: 276855)
- 10% neutral buffered formalin (Sigma-Aldrich, catalog number: HT501128)
- DAPI (Stock concentration 10 mg/ml) (Sigma-Aldrich, catalog number: D9542)
- LIVE/DEAD® Viability/Cytotoxicity Kit (Thermo Fisher Scientific, catalog number: L3224)
- Methanol (Sigma-Aldrich, catalog number: 34860)
- ProLong® Gold Antifade Mountant (Thermo Fisher Scientific, catalog number: P36930)
- Silicon rhodamine far-red fluorophore-conjugated DNA minor groove binder bisbenzimide (SiR-DNA) (tebu-bio Ltd, catalog number: SC007)
- Phenol red-free DMEM with HEPES (Life Technologies, catalog number: 21063029), also referred to as 'Phenol red-free DMEM with HEPES basal media” in the Recipes
- Bovine Serum Albumin (BSA) (Sigma-Aldrich, catalog number: A7030)
- Triton X-100 (Sigma-Aldrich, catalog number: X100)
- (Optional) para-Nitroblebbistatin (Cayman Chemical Company, catalog number: 13891)
- (Optional) Cytochalasin D (Sigma-Aldrich, catalog number: C8273)
- HBSS/HEPES ice cold buffer (see Recipes)
- Agarose solution (see Recipes)
- SF-DMEM (see Recipes)
- Image media (see Recipes)
- MTT working solution (see Recipes)
- 70% ethanol (see Recipes)
- 70% methanol (see Recipes)
Equipment
- Curved dissecting forceps, 10 cm, Serr/C (World Precision Instruments Ltd, catalog number: 15915)
- Fine tip dissecting scissors, 10 cm, straight (World Precision Instruments Ltd, catalog number: 14393)
- Surgical scissors, 14 cm, straight (World Precision Instruments Ltd, catalog number: 14192)
- Spring scissors, 12 cm straight, 12 mm extra-fine blades (World Precision Instruments Ltd, catalog number: 14125)
- Stainless steel blades for vibratome (Campden Instruments LTD, catalog number: 7550-1-SS)
- Automated vibratome (Compresstome® VF-300-0Z; Precisionary Instruments LLC, USA)
- Incubator (Humidified, 37 °C, 5% CO2)
- -20 °C freezer
- GFP filter, excitation 450-490 nm, emission 500-550 nm (for EpCAM-FITC)
- Cy-5 filter, excitation 625-655 nm, emission 665-715 nm (for SiR-DNA and PECAM)
- Plate reader (Tecan; SunriseTM, INSTSUN-1)
- Zeiss Axio Observer inverted widefield microscope, with Lumencor Spectra X LED light source and Hamamatsu Flash 4.0 camera (Zeiss, Germany)
- Zeiss LSM-510 inverted confocal microscope (Zeiss, Germany, model: LSM 510)
- Leica DM2500 widefield microscope (Leica Microsystems, model: Leica DM2500)
Software
- Zen2 acquisition software, blue version (Zeiss, Germany)
- ZEN 2009 (black edition) software (Zeiss, Germany)
- FIJI (ImageJ, version 2.0)
- Icy open source bioimaging analysis software (Version 1.9.8.0; created by the Quantitative Image Analysis Unit at Institut Pasteur, Paris, France)
- Huygens deconvolution software (Scientific Volume Imaging, SVI, Essential version 17.10)
- NIS-Elements (Version 4.50, Nikon Instruments, UK)
- GraphPad Prism version 5
- Microsoft Excel (Microsoft Office 2011 version)
- Microsoft PowerPoint (Microsoft Office 2016 version)
Procedure
- Producing precision-cut lung slices
- Mouse dissection and lung harvesting
- Humanely kill postnatal day 3 (P3), P7, P14 or adult (6-8 weeks) C57BL/6 mice by intraperitoneal injection of pentobarbital (50 mg/kg body weight). Inject 25 µl of pentobarbital for P3, 50 µl for P7, 75 µl for P14 and 100 µl for adult mice using 29 G, 1 ml Insulin Syringes (Figure 1A7). Mice will die in 5-10 min after injection. Confirm death by exsanguination of femoral artery.
- Place the mouse on a dissection tray in supine position. Spray 70% ethanol over the anterior aspect of the abdomen, chest and neck.
- Using curved forceps (Figure 1A2) to pinch the skin from the anterior abdominal wall and gently pull it up, cut a small piece of skin leaving the inner peritoneum intact.
- Using a pair of fine scissors (Figure 1A4) make a longitudinal incision along the midline of anterior abdomen and chest wall and extend it up to the chin (Figure 1B).
- Using two pairs of curved forceps, retract the skin on both sides to expose the anterior aspects of the abdomen, chest wall and neck. (Figure 1Ci) (Video 1).
- Using curved forceps carefully lift the peritoneal wall at the upper part of the abdomen, just below the costal margin and make a small opening using the tip of the fine scissors.
- Extend the opening to the right and left using fine scissors. This will expose the diaphragm.
- Gently lift the lower end of the sternum and make an opening on the upper part of diaphragm using the tip of the fine scissors. Then extend the opening to both side of the diaphragm along the inner surface of the costal margins (Figure 1C i, ii, yellow dotted line). At this stage you will be able to see pale pink colored right and left collapsed lungs (Figure 1C iii, arrow).
- Lift the chest wall by holding the lower end of the sternum with curved forceps and carefully make a parasternal incision (to the left and right) to separate anterior chest wall from the body (Figure 1C i, iii, yellow dotted line) (Video 1).
- Using curved forceps and fine scissors, remove muscle and fascia from the anterior and lateral aspects of the neck wall.
- Using curved forceps, make an opening between the posterior aspect of the trachea and the anterior wall of the esophagus at the midpoint of the neck (Video 1).
- Insert a piece of sterile suture through the opening and place it perpendicular to the trachea at its midpoint (Figure 1).
- Make a small opening in the anterior wall of the trachea just below the cricoid cartilage using fine tipped straight spring scissors (Figure 1A, (6), 1C (iii) yellow arrow, Figure 1D (ii) inset).
- Carefully insert a rigid metallic cannula 25 G for P3, 23 G for P7 and 21 G for P14 and adult mice through the trachea up to a millimeter above the bifurcation of the principal bronchi and fix in place with a suture (Figure 1C (iv) and 1D (ii) inset). The suture was tied, with moderate tightness, around the trachea including cannula inside, by applying two knots.
- After cannulation, inflate the lungs by gently injecting 1.5% low-melting-point agarose prepared with HBSS/HEPES buffer warmed to 37 °C (see Recipes). Use a 5 ml syringe to instill the agarose and keep the syringe attached to the cannula until the agarose solidifies. Inflate both lungs keeping them in situ within the chest cavity using 0.2 ml for P3, 0.275 ml for P7, 0.35 ml for P14 and 1 ml for adult lungs. These volumes enable the lungs to be fully inflated without hyper- or sub-optimal inflation (Figures 1C and 1D).
- After inflation, place a piece of double layered laboratory blue roll over the exposed chest wall and put some wet ice on to the blue roll to solidify the agarose.
Note: Depending on mouse age, this will take 1 to 2 min. - Remove the cannula by gently pulling it out from the trachea and tighten the suture knot.
- Excise the trachea from its upper (mouth) end. Lift the trachea by holding the free ends of the suture and gently remove the entire heart and lungs from the chest cavity (Figures 1C and 1D).
- Immerse the whole heart and lungs in ice-cold serum-free DMEM (SF-DMEM) in a 50 ml centrifuge tube, secure the cap and store on wet ice until slicing. The tissue can be kept on ice for 2-3 h.Video 1. Mouse dissection: from skin incision to trachea mobilization. (All animal maintenance and procedures were carried out according to the requirements of the Animal (Scientific Procedures) Act 1986. Animal work was approved by the South Kensington and St. Mary’s AWERB committee, Imperial College London.)

Figure 1. Mouse dissection and lung harvesting workflow. A. Dissection equipment: 1. Suture, 2. Curved forceps, 3. Fine tip forceps, 4. Fine tip dissecting scissors, 5. Surgical scissors, 6. Spring scissors, 7. Pentobarbital in 1 ml syringe attached with hypodermic needle. B. Skin incision line. C. Adult mouse dissection, agarose inflation and harvesting of lungs. D. Postnatal day 3 (P3) mouse dissection, agarose inflation and harvesting of lungs.
- Precision-cut lung slicing
- Prepare the automated vibratome: Sterilize the buffer tray of the vibratome thoroughly by spraying with 70% ethanol and leave to dry (Figure 2A). Attach vibratome blade to the blade holder using super glue and leave to dry. Sterilize specimen holder and specimen tube using 70% ethanol spray and air dry. Place the syringe chilling block in -20 °C freezer for 15-30 min to cool it down prior to use (Figure 2B).
- Have ready approximately 30 ml of 1.5% agarose gel made with HBSS/HEPES at 37 °C temp. Prepare 200 ml of sterile ice cold 1x HBSS/HEPES buffer and keep on ice. Prepare a 24-well plate with 1 ml ice cold, sterile SF-DMEM per well and keep on ice.
- In a 10 cm diameter Petri dish, separate the left lung lobe from the left bronchus using a scalpel blade (Figures 2C and 2D).
- Excise a tiny section of tissue from the basal end of the lung using the scalpel blade (Figure 2D; outline of the tissue excised is marked with dotted line).
Note: This helps the lobe to sit on the specimen holder vertically. - Place the lung lobe on a piece of clean laboratory blue roll to soak up excess media (Figure 2D).
- Place a tiny drop of super glue onto the cutting end of the specimen holder (Figure 2Ei) and gently place the cut end of the lung lobe onto the glue drop, hold it vertically with curved forceps until the lung lobe tightly adheres to the specimen holder (Figure 2Eii).
Note: It takes 15-30 s to attach the lung lobe onto the specimen holder. - Insert the specimen holder into the metal specimen tube, hold it vertically keeping the lung lobe inside the tube and facing vertically up (Figure 2Eiii).
- Using a 5 ml syringe, fill the specimen tube with 37 °C warm 1.5% agarose so that the entire lobe is submerged in agarose (Figure 2Eiii).
- Place the cold syringe chilling block (Figure 2B) around the specimen tube and hold it in place until the agarose tissue block solidifies (Figure 2Eiv).
Note: It takes about a minute to solidify the agarose. - Insert the specimen tube into the buffer tray and align blade and cutting end of the agarose tissue block.
- Fill the buffer tray with ice cold HBSS/HEPES buffer. Set the desired thickness of tissue slices to 300 µm. Set the cutting speed to number 5 and cutting oscillation frequency to 5. Operate the vibratome in ‘continuous mode’.
- Start collecting transverse precision-cut lung slices (PCLS). Remove the lung slices from the first 1/3rd of the lung lobe from the buffer tray using a small metallic spatula. Collect the slices from the middle 1/3rd of the lobe and place them in the wells of a 24-well plate containing ice cold SF-DMEM (Figure 2F). The middle third of the lung lobe provides almost equal sized slices. Take 1 lung slice in each well in 1 ml media. (Using this technique, a P3 left lung provides 12 slices and an adult lung provides approximately 36 slices.)
Note: It is important to use equal sized slices for imaging and metabolic assays, the remaining slices can be used for additional assays, e.g., immunostaining, protein or RNA extraction. - Incubate PCLS at 37 °C in the presence of 5% CO2 for 2 h and then wash twice with warm SF-DMEM to remove excess agarose from around the tissue in sterile conditions, under the flow hood.
Note: Some agarose will remain in the airspaces within the lungs and this is important to retain lung architecture during culture. - Incubate the PCLS for a further 1 h in SF-DMEM at 37 °C in the presence of 5% CO2. From this point the PCLS are ready for live imaging and further experimentation.
Note: Use the samples immediately for imaging (and migration analysis); but slices can be kept in culture overnight for other experimentation.
- Mouse dissection and lung harvesting
- Live cell imaging of lung alveologenesis
- Label-free, bright-field live imaging
To visualize gross structural changes in lung slices over time, live bright-filed imaging can be performed as follows:- Use phenol red-free DMEM and ibidi 24-well μ-plate for live imaging.
- Prepare transwells for imaging: using scissors cut and remove the rim of the transwell so that the transwell filter comes into contact with the lung slice at the bottom of the well (Figure 2G). Wash the transwells with HBSS (warm) to remove debris and place them in unused wells of ibidi 24-well μ-plate without buffer.
- Add 50 µl of warm (37 °C) phenol red-free SF-DMEM to the center of a well of the ibidi μ-plate.
- Using a thin sterile metallic spatula take a PCLS from Step A2n above and carefully place at the center of the well of ibidi μ-plate. Make sure that the lung slice is placed in the center and is flat and not folded.
- Then gently place the modified transwell on top of the PCLS.
- Add 500 µl phenol red-free SF-DMEM to the upper chamber and 300 µl to the bottom chamber of the transwell.
- To keep the PCLS in place, put a 1.66 g metal flat washer with a central hole on top of the transwell housing (Figures 2H and 2I).
Note: This weight puts light pressure on tissue slice through the permeable transwell membrane, which holds the slice in place without damaging the tissue. The metal flat washer weight should not exceed 3.32 g. - Incubate the PCLS at 37 °C in presence of 5% CO2 and 95% air (~21% O2) in a humidified incubator for 2 h. This incubation step allows the lung slice to settle down prior to image acquisition.
Note: Similar sized PCLS should be allocated for MTT viability assay after live images, as shown in Figure 3. - At the same time, humidify and pre-equilibrate the incubator chamber of an inverted Zeiss Axio Observer widefield epifluorescence microscope (or similar) for 30 min to 1 h with the following conditions: 37 °C, 5% CO2 and room air oxygen levels, approx. 21%.
- After incubation and pre-equilibration transfer the ibidi 24-well plate containing PCLS to the microscope incubator and maintain the incubator chamber conditions as above throughout the entire time-lapse duration.
- Start capturing time-lapse images using a long working distance 40x (0.7 NA, air) objective lens under bright-light (Figure 2I).
- For bright-field imaging, find the best focused plane at the region of interest and keeping this point as the center select 30 z-slices towards the upper surface of the slice and 30 slices towards the bottom surface of the slice with 1 µm step-gap between the slices (a total of 60 slices in the middle 60 µm z-axis of tissue).
- Set the time-lapse image capture interval to every 15 min for 12-19 h.
Note: Four images per hour (15 min interval) reduces light exposure, hence minimizes fluorophore bleaching but produces smooth time-lapse video with 10-12fps (see, Supplementary Movie 2 in Akram et al, 2019). - Select 4 separate fields of interest within alveolar regions, avoiding airways, from the four quadrants of a PCLS.
Note: The 2 h incubation and pre-equilibration of the microscope incubator are crucially important to avoid the plane of focus drifting during time-lapse image acquisition. Image 4 separate PCLS per experiment.
Figure 2. Precision-cut lung slicing and live imaging setup. A. Lung slicer: Vibratome, Compresstome® VF-300-0Z. B. Syringe chilling block for solidifying agarose. C. Agarose inflated lungs harvested from a P3 mouse. D. Separated left lung lobe on laboratory tissue roll. E. Schematic diagram showing agarose embedding of left lung lobe on specimen holder for making PCLS. F. Precision-cut lung slices are in 24-well plate. G. Modification of transwell for stabilizing lung slices on well plate. H. Precision-cut lung slices are set in ibidi 24-well μ-plate using modified transwells and metal flat washer (black rim). I. Schematic showing the relative position and alignment of metal flat washer, transwell, lung slice, and 40x long working distance air objective lens on an inverted microscope for live imaging.
- Live fluorescence imaging of immunostained PCLS
Dual staining of PCLS for live imaging- To visualize and track epithelial cells within the PCLS, dual stain cells with FITC-conjugated EpCAM antibody (EpCAM-FITC) and SiR-DNA. (EpCAM selectively labels epithelial cell membranes. SiR-DNA is silicon rhodamine far-red fluorophore-conjugated DNA minor groove binder bisbenzimide and stains the nuclei of cells).
- Incubate PCLS (from Step A2n) for 1 h at 37 °C with EpCAM-FITC antibody diluted 1:200 and SiR-DNA diluted 1:300 in DMEM (without shaking). Use 500 µl of antibody/SF-DMEM media for each well per PCLS.
- To visualize epithelial and endothelial cells within the alveolar region, incubate PCLS for 1 h at 37 °C with dual Alexa-647 conjugated PECAM antibody and EpCAM-FITC antibody both at 1:200 in 500 µl SF-DMEM per sample per well.
- After incubation, wash the PCLS three times with warm SF-DMEM.
- Image media: Prepare image media by adding EpCAM-FITC antibody at 1:500 and SiR-DNA at 1:1000 for epithelial cells or PECAM at 1:500 along with EpCAM-FITC (1:500) for dual-labeling of epithelial/endothelial cells in 800 µl phenol red-free SF-DMEM for each PCLS per well.
Note: SiR-DNA at 1:1000 dilution can be used as a nuclear marker for cells that are dual labeled with epithelial or endothelial markers. - Place a 50 µl drop of image media at the center of a well in an ibidi 24-well μ-plate and carefully place a dual stained PCLS onto the image media using a thin metallic spatula.
- Place a modified transwell on top of the PCLS, add 500 µl of image media to the upper chamber and 300 µl image media to the bottom chamber and finally stabilize the transwell by placing a flat metal washer onto the transwell as described above (Figures 2H and 2I).
- Incubate the PCLS at 37 °C in presence of 5% CO2 and 95% air in a humidified incubator for 2 h. This incubation step intensifies the staining and allows the lung slice to settle down prior to image acquisition.
- Pre-equilibrate the microscope incubator chamber as described for bright-field imaging (as Step B1i).
- After incubation and pre-equilibration transfer the 24-well plate to an inverted Zeiss Axio Observer widefield epifluorescence microscope incubator and maintain the incubator conditions as above throughout the time-lapse duration.
- Start recording time-lapse images using a long working distance 40x (0.7 NA, air) objective lens using a GFP filter, excitation 450-490 nm, emission 500-550 nm (for EpCAM-FITC) and Cy-5, excitation 625-655 nm, emission 665-715 nm (for SiR-DNA and PECAM) from 4 fields of alveolar regions per slice for 8-19 h at 15 min intervals. Capture 11 images from the middle 11 µm thickness of tissue along the z-axis with a 1 µm step-gap between each slice to make a z-stack from each PCLS.
Notes:- Perform time-lapse imaging on four PCLS at a time per experiment. Do not capture bright-field images and fluorescent images from the same PCLS within an experiment. Exposure of bright light significantly bleaches the intensity of fluorescent staining. Do not use a UV or DAPI filter (excitation 358 nm and emission 463 nm) for live imaging of PCLS as this causes significant cell death. Zen2 acquisition software, blue version was used for both brightfield and fluorescence live imaging.
- During live cell imaging, cellular dynamics or other behaviors can be manipulated by adding various factors such as cytokines or small molecules inhibitors into the image media. The resulting cell responses can be recorded in real-time. For example, we recently added the actin cytoskeleton modifiers Blebbistatin and Cytochalasin-D to PCLS and assessed their effects on cell migration using this live-PCLS imaging technique (Akram et al., 2019).
- To ensure the viability of PCLS, perform a viability assay on PCLS after every time-lapse live imaging experiment. For bright-field imaging on unlabeled PCLS perform Live/Dead staining and for fluorescence imaging on dual stained PCLS, perform an MTT assay.
- Duration of experiment p: From mouse dissection to the start of time-lapse imaging takes 7-8 h.
- Label-free, bright-field live imaging
- Cell viability assays on PCLS after live cell imaging experiments
- MTT assay
- Place four PCLS that have undergone live imaging into an ibidi 24-well μ-plate, 1 slice per well. Place four similar sized, non-imaged PCLS that have been kept in an incubator with 37 °C, 5% CO2 for the same duration as the imaged slices in a conventional 24-well plate, 1 slice per well in phenol red-free SF-DMEM. Four similar-sized slices were selected to perform MTT assay to determine cell viability at the start of time-lapse imaging experiments (Figure 3).
- To assess metabolic activity within cells at the initiation of live imaging (0 h control), add 500 µl of 10% MTT solution (Stock Con. 5 mg/ml) made with SF-DMEM to each well.
Note: This MTT needs to be done before starting live imaging (Figure 3B). - Incubate the PCLS at 37 °C in presence of 5% CO2 for 1 h (Figure 3C).
- Remove the MTT media from the wells.
- Solubilize the formazan crystals formed within the viable cells by adding an equal volume of DMSO (500 µl) and incubating at 37 °C for 10 min.
- Take 200 µl of eluted formazan solution from each PCLS and place into individual wells of a 96-well plate (Figure 3D).
- Measure the absorbance (OD) at 570 nm and correct at 690 nm using a plate reader.
- To assess metabolic activity within post-time lapse PCLS, carefully remove the transwells after completion of time-lapse imaging. If PCLS adhere to the under surface of the transwell PET membrane, flush with warm HBSS using a pipette, to detach it.
- Wash the PCLS with warm HBSS once.
- Then perform MTT assay on PCLS undergone time-lapse imaging and non-imaged PCLS cultured in a conventional incubator in the same way as described for initial time-point control PCLS above.
- Compare the initial time-point (0 h) and post-time lapse (Imaged and non-imaged PCLS) OD values to evaluate cell viability after time-lapse imaging (For examples, see Figure 1 j in reference Akram et al, 2019).

Figure 3. MTT assay workflow. A. Allocation of PCLS for MTT assay. B. Plating of PCLS in 24-well plates for treating with MTT solution. C. PCLS are treated with MTT solution. D. Eluted formazan in wells of a 96-well plate.
- Live/Dead assay
- Perform cell viability assay on PCLS using LIVE/DEAD® Viability/Cytotoxicity Kit.
- Place four PCLS into an ibidi 24-well μ-plate for time-lapse imaging and four PCLS into a conventional incubator in a 24-well plate to culture for the same duration as the time-lapse without imaging, in 500 µl phenol red-free SF-DMEM. Place four PCLS into an ordinary 24-well plate to perform Live/Dead assay to evaluate viability at the initial time-point (0 h).
- For the non-imaged slices initial time-point (0 h), remove phenol red-free SF-DMEM and incubate PCLS with 2 µM Calcein AM and 2 µM Ethidium homodimer-1 (EthD-1) in 250 µl warm HBSS for 30 min at 37 °C, 1 PCLS per well.
- Wash the PCLS two times with HBSS and fix with 10% neutral buffered formalin for 30 min at room temperature (RT). Wash twice with HBSS and store the slices in HBSS at 4 °C prior to imaging.
- At the end of time-lapse imaging, remove transwells from each well as described for MTT assay and wash once with HBSS. Then perform Live/Dead assay on both time-lapse imaged and non-imaged PCLS, i.e., those cultured in conventional incubator for same duration as time-lapse, as above.
- As a positive control for dead cells, treat PCLS with 70% methanol for 30 min at RT and incubate with Calcein AM and EthD-1 as described above.
- Mount the PCLS on glass slides with a drop of ProLong® Gold Antifade Mountant and cover with a glass coverslip. Allow 1 h for mountant to set at room temperature.
- Capture images using a Zeiss LSM-510 confocal microscope with a 20x 0.8 NA air objective and ZEN 2009 (black edition) software (or equivalent).
- MTT assay
- Cell tracking for cell migration quantification using Icy software
- Open the Icy software and import an EpCAM-FITC, SiR-DNA labeled raw time-lapse image sequence file.
- In the FITC channel, select the best focused plane, i.e., a single slice from the z-stack of the image sequence (Figure 4B).
- Select the desired time duration you wish to conduct cell tracking for (e.g., 8-16 h).
- Use the Spot detection and Tracking tools (under the Detection/Tracking tab) to detect EpCAM positive epithelial cells and track them throughout the pre-defined time-lapse duration.
- To detect EpCAM-FITC positive cells, set the Spot detector parameters as follows on full frame image: (a) Detect bright spot over dark background option, (b) Size of spots 7 and 13 pixels with 100% sensitivity, (c) Filtering parameter with 100-200 (adjust accordingly to correctly spot the individual cell), (d) Enable swimming pool option under the Output tab. Then Click ‘Start detection’. Check that the detector is identifying individual cells (which will be marked by red circle). If necessary correct spot detection by altering the Pixel size and Filtering parameters.
- After optimization of spot detection, click ‘Run Tracking’ to quantify the cell migration. Quantify cell migration on X-Y axis only.
Note: 3D X-Y-Z axis migration quantification does not work accurately under this setting. - After tracking, check every cell migration trail in each image field by rewinding the video sequence on Tracking mode a few times to see if there is any off-target tracking. If there is any off-target tracking, deselect these trails. Off-target tracking can occur when a trail is falsely generated without the presence of a migrating cell (Figure 4B).
Note: This manual checking is crucial as under this setting Icy often tracks off-target objects. For cell tracking do not perform batch processing, it does not work under this setting. Track cells of each field from each PCLS individually slice by slice and by adjusting the detection parameters with post-tracking off-target screening for each video sequence. - After completion of tracking add ‘Motion profiler processor’ to the ‘Track processor’ option under Track manager. Use Real unit option (µm/s) for migration and speed quantification from Track manager (Figure 4A).
- Export data as Excel file and the cell tracking trace as image file (Figures 4A and 4C).
Note: For an example of generated cell tracking data, and cell migration data, see Figures 2 h, i, j in the reference Akram et al, 2019.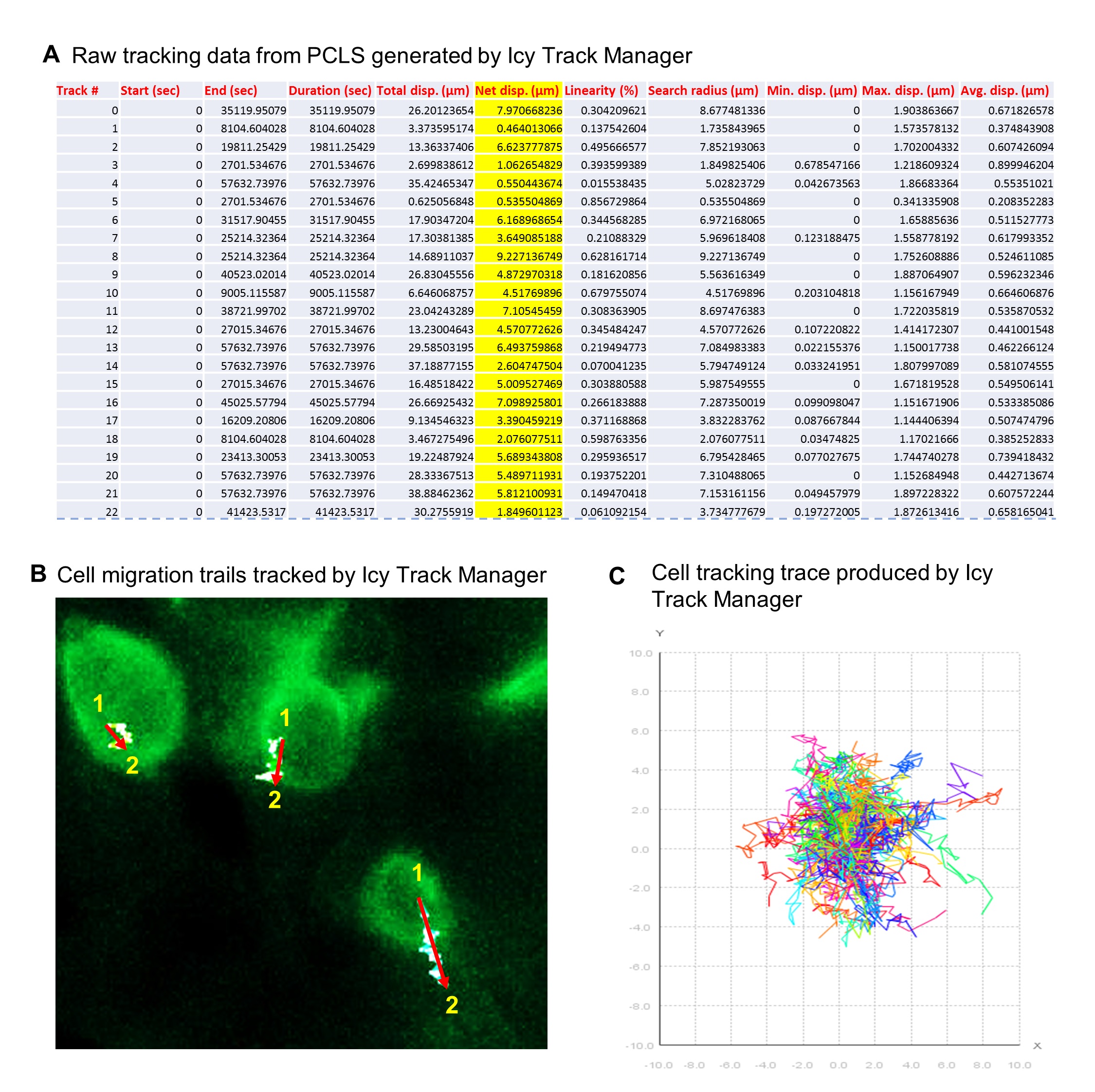
Figure 4. Cell tracking by Icy. A. Raw cell tracking data from P3 PCLS generated by Icy Track Manager (22 out of 118 cell tracking data presented). B. Cell migration trails of 3 EpCAM-FITC positive cells from P3 PCLS tracked by Icy Track Manager showing 3 cells migrated from position 1 to position 2 in 16 h (zoomed raw image). Red arrows indicate the linear ‘Net cell migration’. C. Cell tracking trace produced from 1 full-frame visual filed from a P3 PCLS by Icy Track Manager.
- Image deconvolution and movie generation
- Image deconvolution using Huygens deconvolution software
Note: The raw fluorescent images obtained by widefield microscopy from 300 µm thick PCLS are hazy and unsuitable for individual cell observations due to the limitations of widefield imaging, and light diffraction caused by sample thickness. Deconvolution software uses an algorithm to eliminate out-of-focus light and produce sharper images. (See Figure 1 i and Supplementary movie 2 in reference Akram et al., 2019).- Upload the widefield z-stack of time-lapse fluorescent images to the Huygens deconvolution software (Step B2k). Each z-stack consists of 11 separate z-slices per sample, with a 1 µm step between slices.
Note: The optimum step-gap between slices is dependent on the objective lens and requirements of the deconvolution software. For Huygens deconvolution software for 40x objective lens optimum step-gap is no more than 1 µm. This configuration may vary for other deconvolution software packages. Optimizing the step-gap is crucial to generate confocal-like, deconvolved images. - Deconvolve the image sequence using the built-in automated CMLE algorithm with the following parameters: (a) Maximum iterations 50, (b) Signal to noise ratio 40, (c) Quality threshold 0.01, (d) Iteration mode Optimized.
- Save the deconvolved time-lapse image sequence as a TIFF z-stack image sequence file for 2D and 3D time-lapse video generation.
- Upload the widefield z-stack of time-lapse fluorescent images to the Huygens deconvolution software (Step B2k). Each z-stack consists of 11 separate z-slices per sample, with a 1 µm step between slices.
- 2D and 3D and bright-field video generation
- For 2D video generation, import deconvolved, time-lapse z-stacks into Icy software (Step E1c). Select the best-focused single z-plane, adjust the brightness and contrast and crop the area of interest. Save the file as .avi file format.
- For 3D video reconstruction, import deconvolved z-stack time-lapse files into NIS-Elements and align to correct for X-Y drift (Step E1c). Generate 3D videos using the ‘Volume View’ and ‘Movie Maker’ modules of NIS-Elements.
- For 2D brightfield video generation, import raw image z-tack files into Fiji (ImageJ) software. Select the best-focused single z-plane, adjust brightness and contrast. Save the file as .avi video file.
Note: Brightfield images do not need deconvolution. - For annotation on 2D fluorescent and brightfield videos, insert the .avi video files on Microsoft PowerPoint (MS Office 2016) as video file, resize the slide size to match with the video window dimensions, annotate the video accordingly using Text and Shape tools. Export the PowerPoint file as high resolution .mp4 movie file. (See supplementary movies in Akram et al., 2019).
- Image deconvolution using Huygens deconvolution software
Data analysis
- Quantification of cell behaviors
- To quantify cellular events, such as cell clustering, hollowing, septation and cell extension during post-natal alveologenesis in PCLS carefully examine deconvolved and brightfield time-lapse video files (using 40x objective) on screen. Record the number of times each event is observed in a single field of view from a video file. Examine videos from at least 4 different fields from different PCLS and from 3 independent experiments.
- Present data as the frequency of each type of cellular behavior as the total number of observations of that behavior per field. (For examples of results, see Results section, pages 4-7 in Akram et al., 2019)
- Quantification of cell migration
- Track manager in Icy produces a range of cell migration information from the tracking data including: Net cell migration, Total displacement, migration speed (Figure 4A).
- To quantify and compare epithelial cell migration between different postnatal age groups (e.g., P3, P7, P14 and Adult) or different treatment conditions determine (a) Net cell migration and (b) the net distance traveled by individual cells within a specified time-period. Net cell migration is the mean value calculated to present how much linear distance in the X-Y axis (Initial point A to end point B) a cell migrates. A mean value of migrated distances of the total number of cells from each field is presented as mean net cell migration. (For example of results, see Figure 2 h, i, j in Akram et al., 2019).
Note: For migration analysis use ‘Net Displacement’ not ‘Total Displacement’ data from cell tracking record (Figure 4A, yellow highlight). - There are two groups of epithelial cells present in the lung slices: one group of cells are sessile, and other group of cells are highly motile. To differentiate these two groups and to compare them between different postnatal age groups and treatment conditions, determine the net distance traveled by individual cells. Rank them and present as percent or proportion of cells that migrate the ranked distances (See Figure 2 h-j and Figure 6 k-n in Akram et al., 2019).
Recipes
- HBSS/HEPES ice cold buffer
1x HBSS
1% HEPES 1 M - Agarose solution
1.5 g low-melting point agarose powder
100 ml HBSS/HEPES buffer
Dissolve agarose in HBSS/HEPES buffer in microwave. Prepare 30 ml agarose for processing 1 lung - SF-DMEM
DMEM basal media
1% Penicillin-Streptomycin - Image media
Phenol red-free DMEM with HEPES basal media
1% Penicillin-Streptomycin
EpCAM-FITC antibody 1:500
PECAM antibody 1:500 or SiR-DNA 1:1,000 - MTT working solution
DMEM basal media
10% MTT stock solution (Stock Con. 5 mg/ml in DMSO)
Note: Stock solution is stable for 1 year when stored at -20 °C in a dark container. - 70% ethanol
70 ml of absolute ethanol
30 ml distilled water - 70% methanol
70 ml methanol
30 ml PBS
Acknowledgments
This project was funded by a Leverhulme Trust grant to CHD (RPG-2015-226). The Facility for Imaging by Light Microscopy (FILM) at Imperial College London is part-supported by funding from the Wellcome Trust (grant 104931/Z/14/Z) and BBSRC (grant BB/L015129/1). This protocol is adapted from the method published in Akram et al., 2019.
Competing interests
The authors declare that they have no conflict of interest.
Ethics
All animal maintenance and procedures were carried out according to the requirements of the Animal (Scientific Procedures) Act 1986. Animal work was approved by the South Kensington and St. Mary’s AWERB committee, Imperial College London.
References
- Akram, K. M., Patel, N., Spiteri, M. A. and Forsyth, N. R. (2016). Lung regeneration: endogenous and exogenous stem cell mediated therapeutic approaches. Int J Mol Sci 17(1).
- Akram, K. M., Yates, L. L., Mongey, R., Rothery, S., Gaboriau, D. C. A., Sanderson, J., Hind, M., Griffiths, M. and Dean, C. H. (2019). Live imaging of alveologenesis in precision-cut lung slices reveals dynamic epithelial cell behaviour. Nat Commun 10(1): 1178.
- Amy, R. W., Bowes, D., Burri, P. H., Haines, J. and Thurlbeck, W. M. (1977). Postnatal growth of the mouse lung. J Anat 124(Pt 1): 131-151.
- Burgstaller, G., Vierkotten, S., Lindner, M., Konigshoff, M. and Eickelberg, O. (2015). Multidimensional immunolabeling and 4D time-lapse imaging of vital ex vivo lung tissue. Am J Physiol Lung Cell Mol Physiol 309(4): L323-332.
- Herriges, M. and Morrisey, E. E. (2014). Lung development: orchestrating the generation and regeneration of a complex organ. Development 141(3): 502-513.
- Hilgendorff, A., Reiss, I., Ehrhardt, H., Eickelberg, O. and Alvira, C. M. (2014). Chronic lung disease in the preterm infant. Lessons learned from animal models. Am J Respir Cell Mol Biol 50(2): 233-245.
- Hind, M., Corcoran, J. and Maden, M. (2002). Alveolar proliferation, retinoid synthesizing enzymes, and endogenous retinoids in the postnatal mouse lung. Different roles for Aldh-1 and Raldh-2. Am J Respir Cell Mol Biol 26(1): 67-73.
- Kotton, D. N. and Morrisey, E. E. (2014). Lung regeneration: mechanisms, applications and emerging stem cell populations. Nat Med 20(8): 822-832.
- Kreiger, P. A., Ruchelli, E. D., Mahboubi, S., Hedrick, H., Scott Adzick, N. and Russo, P. A. (2006). Fetal pulmonary malformations: defining histopathology. Am J Surg Pathol 30(5): 643-649.
- Li, J., Wang, Z., Chu, Q., Jiang, K., Li, J. and Tang, N. (2018). The strength of mechanical forces determines the differentiation of alveolar epithelial cells. Dev Cell 44(3): 297-312 e5.
- Madurga, A., Mizikova, I., Ruiz-Camp, J. and Morty, R. E. (2013). Recent advances in late lung development and the pathogenesis of bronchopulmonary dysplasia. Am J Physiol Lung Cell Mol Physiol 305(12): L893-905.
- McGowan, S. E. (2014). Paracrine cellular and extracellular matrix interactions with mesenchymal progenitors during pulmonary alveolar septation. Birth Defects Res A Clin Mol Teratol 100(3): 227-239.
- Meng, Q. J., McMaster, A., Beesley, S., Lu, W. Q., Gibbs, J., Parks, D., Collins, J., Farrow, S., Donn, R., Ray, D. and Loudon, A. (2008). Ligand modulation of REV-ERBalpha function resets the peripheral circadian clock in a phasic manner. J Cell Sci 121(Pt 21): 3629-3635.
- Mund, S. I., Stampanoni, M. and Schittny, J. C. (2008). Developmental alveolarization of the mouse lung. Dev Dyn 237(8): 2108-2116.
- Pieretti, A. C., Ahmed, A. M., Roberts, J. D., Jr. and Kelleher, C. M. (2014). A novel in vitro model to study alveologenesis. Am J Respir Cell Mol Biol 50(2): 459-469.
- Poobalasingam, T., Salman, D., Li, H., Alcada, J. and Dean, C. H. (2017). Imaging the lung: the old ways and the new. Histol Histopathol 32(4): 325-337.
- Roth-Kleiner, M. and Post, M. (2005). Similarities and dissimilarities of branching and septation during lung development. Pediatr Pulmonol 40(2): 113-134.
- Sanderson, M. J. (2011). Exploring lung physiology in health and disease with lung slices. Pulm Pharmacol Ther 24(5): 452-465.
- Snoeck, H. W. (2015). Modeling human lung development and disease using pluripotent stem cells. Development 142(1): 13-16.
- Thornton, E. E., Krummel, M. F. and Looney, M. R. (2012). Live imaging of the lung. Curr Protoc Cytom Chapter 12: Unit12 28.
- Warburton, D., Gauldie, J., Bellusci, S. and Shi, W. (2006). Lung development and susceptibility to chronic obstructive pulmonary disease. Proc Am Thorac Soc 3(8): 668-672.
Article Information
Copyright
© 2019 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Akram, K. M., Yates, L. L., Mongey, R., Rothery, S., Gaboriau, D. C. A., Sanderson, J., Hind, M., Griffiths, M. and Dean, C. H. (2019). Time-lapse Imaging of Alveologenesis in Mouse Precision-cut Lung Slices. Bio-protocol 9(20): e3403. DOI: 10.21769/BioProtoc.3403.
Category
Cell Biology > Cell imaging > Widefield microscopy
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










