- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Experimental Setup for a Diffusion Bioreactor to Isolate Unculturable Soil Bacteria
Published: Vol 9, Iss 19, Oct 5, 2019 DOI: 10.21769/BioProtoc.3388 Views: 6210
Reviewed by: Alba BlesaCristina Colomer-WinterWenrong He

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Plasmodium cynomolgi Berok Growth Inhibition Assay by Thiol-reactive Probe Based Flow Cytometric Measurement
Jessica Jie Ying Ong [...] Jin-Hee Han
Sep 5, 2021 3452 Views

A Microfluidic Platform for Tracking Individual Cell Dynamics during an Unperturbed Nutrients Exhaustion
Théo Aspert [...] Gilles Charvin
Jul 20, 2022 2890 Views

Babesia duncani in Culture and in Mouse (ICIM) Model for the Advancement of Babesia Biology, Pathogenesis and Therapy
Vandana Kumari [...] Choukri Ben Mamoun
Nov 20, 2022 1929 Views
Abstract
Unculturable bacteria are those bacteria which proliferate in their native habitat but unable to grow or thrive in the normal laboratory media and conditions. The molecular techniques have revealed the significance of these uncultured bacteria in terms of their functional diversity and potential to produce secondary metabolites. To achieve these benefits, scientists have attempted to isolate and cultivate unculturable bacteria in the laboratory using transwell plates, optical tweezers, laser microdissection, microbioreactors, and diffusions bioreactors. However, these techniques are still inadequate to resolve the difficulties of cultivating unculturable bacteria. Therefore, it is essential to develop new cultivation method that enables growth of diverse range of bacteria in the laboratory conditions. Diffusion bioreactor is a membrane bound chamber which allows microbes to proliferate in their native environment by providing the excess to naturally occurring nutrients and signaling compounds. This paper presents efficient and reliable protocol to construct a diffusion bioreactor and its utilization to isolate and cultivate unculturable soil bacteria in laboratory.
Keywords: Diffusion bioreactorBackground
The metagenomic analysis has shown an overwhelming number of bacterial populations which are beyond the cultivation in the normal laboratory conditions (Gans et al., 2005; Pham and Kim, 2012). These bacteria which are not-yet cultured and not able to grow on synthetic media are known as unculturable bacteria. The “unculturable” does not imply that these bacteria can never be cultured, but instead, it indicates the absence of information on their natural habitat and growth requirements (Stewart, 2012; Chaudhary et al., 2019). The molecular analysis has reported that uncultured bacteria possess novel biological and chemical properties, produce secondary metabolites, and comprise a diverse functional role (Ferrari et al., 2005; Vartoukian et al., 2010; van Dorst et al., 2016). The secondary metabolites such as novel antibiotics, enzymes, toxins, alkaloids have potential use for human welfares. Due to these beneficial aspects, several studies have attempted to explore and cultivate the hidden uncultured bacteria from the environments (Sait et al., 2002; Ferrari et al., 2005; Davis et al., 2005; George et al., 2011; van Dorst et al., 2016).
There are several difficulties in cultivating unculturable species in the laboratory setups. The unculturable bacteria fail to replicate in laboratory conditions due to lack of natural growth environment and inappropriate incubation time, temperature, and nutrients (Stewart, 2012). Next, traditional approach of cultivation is mainly based on nutrient-rich media which favors only rapid growers (Chaudhary et al., 2019). In the natural habitat, microbe-microbe interaction and their metabolism are crucial aspects for survival and growth. These microbial behaviors depend on several physiological parameters which include pH, osmotic conditions, temperature, and nutrients (Sait et al., 2002). Therefore, without addressing above-mentioned limitations, it is not possible to cultivate missing bacterial species. Strategies such as simulating natural habitat, modifying nutrients composition, optimizing growth conditions, co-culturing with helper agents, and prolonging the incubation time during cultivation of uncultured domains have reported good results (Kaberlein et al., 2002; Joseph et al., 2003; Pham et al., 2012; Stewart, 2012; Chaudhary et al., 2019). Some recent advancement has been made in cultivation techniques using diffusion chambers, transwell plates, microbioreactors, optical tweezers and laser microdissection (Pham et al., 2012). However, the available techniques are still inadequate to address the limitation of cultivation of unculturable bacteria. Any attempt to develop novel cultivation technique for unculturable bacteria accounts a great significance in this field.
Enrichment cultivation utilizing diffusion bioreactor is a promising strategy to cultivate previously uncultured microbes. It helps to enrich microbial species by creating natural environment and allows the microbes with the access to growth components and signaling compounds (Bollmann et al., 2007). In addition, it also provides environment for bacterial interaction (Stewart, 2012; Chaudhary et al., 2019). Diffusion bioreactors are also suitable to cultivate various microbial species from varieties of samples such as soil, sediments, and water samples (Kakumanu et al., 2012; Chaudhary et al., 2019). This paper describes a novel cultivation technique for uncultured soil bacteria using newly designed diffusion bioreactor. Diffusion bioreactor permits the passage of essential elements from the soil into the inner chamber mimicking the natural environments (Kaberlein et al., 2002; Bollmann et al., 2007; Kakumanu and Williams, 2012). Here, we present an effective protocol for experimental setup using diffusion bioreactors to isolate and cultivate the unculturable soil bacteria.
Materials and Reagents
- 2- mm mesh sieve (Fisher Scientific, catalog number: 04-881-10G)
- Whatman filter paper, No. 1 (GE Healthcare, catalog number: 1001-150)
- Mixed cellulose ester filter membrane, 0.22 µm (Merck Millipore, catalog number: GSWP04700)
- Polycarbonate membrane, 0.45 µm (GE Healthcare, catalog number: RPN303B)
- 2 L Conical flasks (DURAN, catalog number: 212176303)
- Petri dish (SPL Life Science, catalog number: 10090)
- 15 ml sterile tube (SPL Life Science, catalog number: 50015)
- Inoculating loop (SPL Life Science, catalog number: 90010)
- Plastic containers with sizes of 2 L (for inner chamber) and 4 L (for outer chamber) (Berry Plastic, catalog number: T51368CP)
- R2A broth (KisanBio, catalog number: MB-R2230)
- Nutrient broth (NB, KisanBio, catalog number: MB-N1037)
- Tryptic Soy Broth (TSB, KisanBio, catalog number: MB-T1053)
- Luria-Bertani broth (LB, Oxoid, catalog number: CM0996)
- Agar (Bio Basic, catalog number: MD01)
- Gelatin (Sigma-Aldrich, catalog number: G2500)
- Cycloheximide (Sigma-Aldrich, catalog number: C7698)
- DNA isolation kit (Instagene Matrix, Bio-Rad, catalog number: 7326030)
- Multiscreen PCR384 Filter Plate kit (Millipore, catalog number: S384PCR10)
- BigDye Terminator Cycle Sequencing Kit (Applied Biosystems, catalog number: 4337455)
- FastDNA SPIN kit for soil (MP Biomedicals, catalog number: 116560-200)
- QIAQuick PCR Purification Kit (Qiagen, catalog number: 28104)
- Beef extract
- Casein
- Enzymatic digest of soya bean
- Glucose
- Peptone
- Sodium pyruvate
- Soluble starch
- Yeast extract
- NaCl
- MgSO4·7H2O
- CaCo3
- K2HPO4
- KNO3
- FeCl2·7H2O
- (NH2)2HPO4
- HCl
- CoCl2·6H2O
- CuCl2·2H2O
- FeCl2·4H2O
- NaBO3
- MnCl2·4H2O
- Na2MoO4·2H2O
- NiCl2·6H2O
- ZnCl2
- NaOH
- Na2SeO3·5H2O
- Na2WO4·2H2O
- Soil extract (SE) (see Recipes)
- Trace element SL-10 (see Recipes)
- Selenite tugstate (see Recipes)
Equipment
- Orbital shaker (FINEPCR, model: SH30)
- Vortex (Scientific Industries, model: 2E-179213)
- Multi-room incubator (Han Baek Scientific, model: HB-103-4)
- Twin cooling system Refrigerator (4 °C to -18 °C) (Samsung Zipel, model: C23442BS300886)
- Centrifuge (Hanil Science Industrial, model: UNION 32R)
- Magnetic stirrer (Global Lab, model: GLMS-G)
- MaestroNano spectrophotometer (MaestroGen, model: MN-913)
- MultiGene OptiMax Thermal Cycler (Labnet International, catalog: TC9600-G-230V)
- 3730xl DNA analyzer (Applied Biosystems, catalog: A41046)
Software
- Microsoft Office Excel 2013 and OriginPro 8.5
- MEGA 6.0.6 (For phylogenetic analysis)
- Lasergene SeqMan Pro, version 7.1.0 (44.1) (DNASTAR Inc.)
Procedure
- Soil sampling
- Collect soil samples (4 kg per experimental set-up) from the uppermost layer (0-10 cm deep) in a sterile zippered bag.
- Sieve the collected soil samples using a 2-mm mesh sieve.
- Utilize the sieved soil for DNA extraction, cultivation, and preparation of soil extract (SE).
- Preparation of culture media
- Prepare R2A, TSA, LB, and NB culture media according to the instructions provided by the manufacturer (Table 1).
- Prepare 50% diluted R2A by mixing equal amount of normal R2A media and distilled water (d/w) (For example 1 L normal R2A media + 1 L d/w).
- Prepare R2A-SE by using SE instead of d/w. (Recipe 1).
- Prepare starch-casein agar-SE (SCA-SE) and J26-SE media by using SE and the composition as mentioned in Table 1 (see Recipe 2). J26-SE media is laboratory formulated media and refer to Table 1 for its preparation.
- Construction of the diffusion bioreactor
- Take 2 L plastic container to prepare it as the inner chamber of the diffusion bioreactor.
- Make holes (6 mm in diameter) in the wall of the inner chamber as shown in Figure 1. To make holes, use electric drill machine or screw driver (with heating) of defined size. Maintain the space of 8 mm in length between the holes.
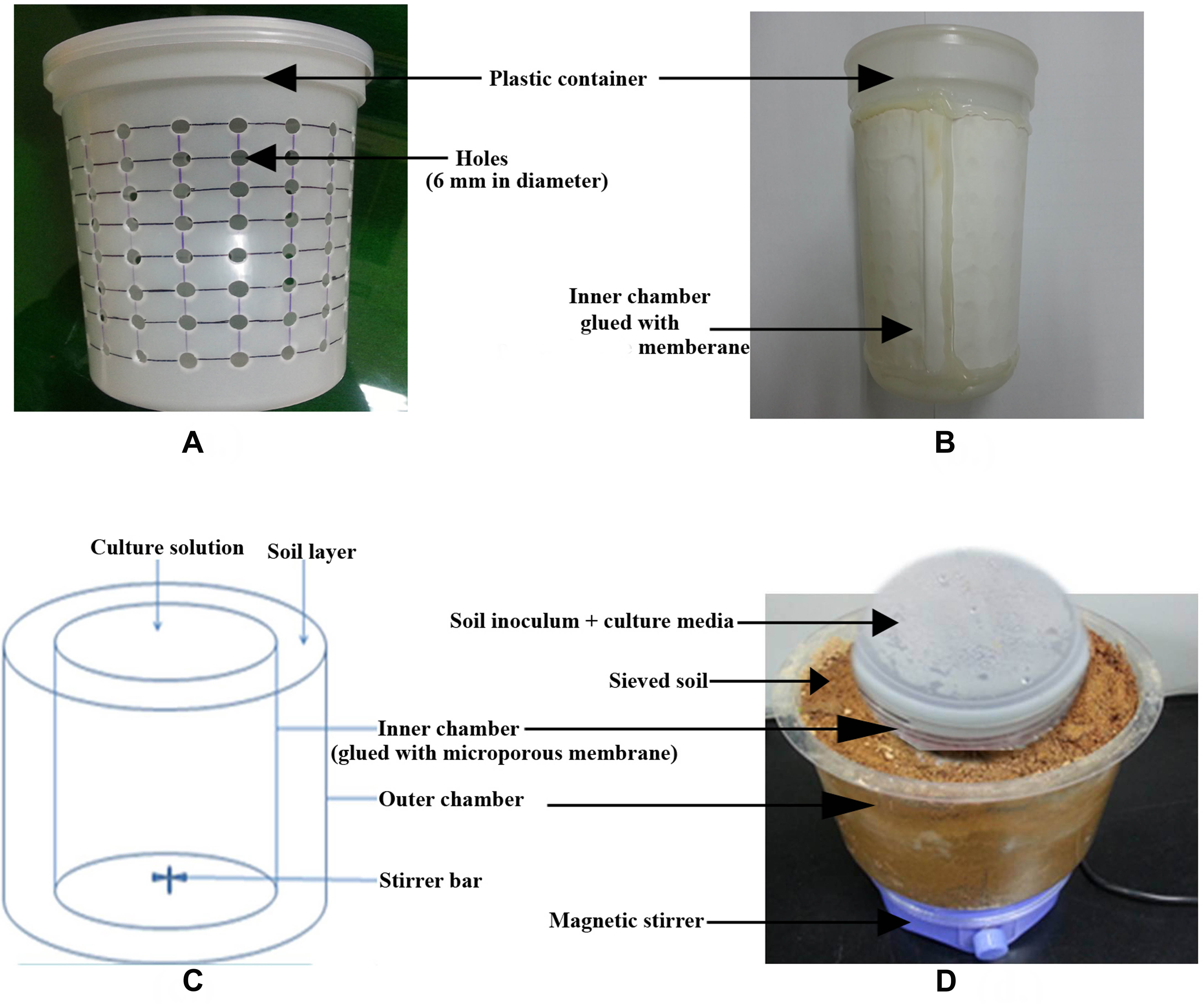
Figure 1. Design of the diffusion bioreactor. A. Plastic container (2 L in size) perforated throughout with holes; B. Inner chamber (2L in size) glued with polycarbonate membrane; C. Schematic representation of the diffusion bioreactor; D. Overall experimental setup using the newly developed diffusion bioreactor (Figure adapted from Chaudhary et al., 2019). - Stick polycarbonate membrane (0.45 µm pore size) on the outer side of the inner chamber.
- Experimental setup with constructed diffusion bioreactor
- Place the inner chamber inside the outer chamber (4 L plastic container).
- Fill the gap between the walls of the two chambers with soil.
- Mix 3 g of soil sample with 300 ml of respective culture medium properly and then add this mixture in the inner chamber. Also, place a magnetic stir inside the inner chamber. (Figures 1 and 2).
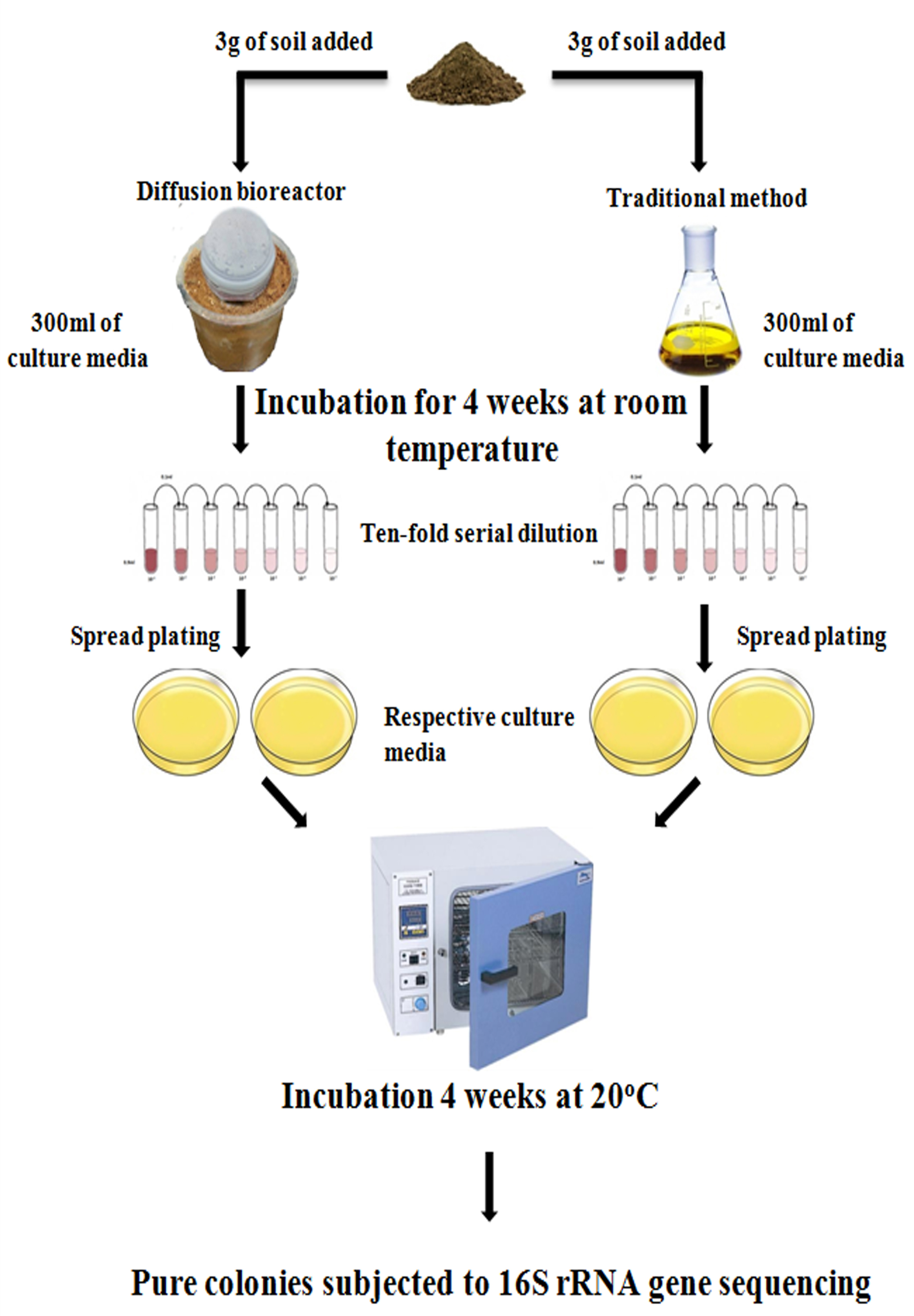
Figure 2. Experimental setup for diffusion bioreactor (Chaudhary et al., 2019) - Close and seal tightly the lid of the inner chamber with the sealing tape.
- Keep stirring (at 60-80 rpm) the diffusion bioreactors on the magnetic stirrer and incubate at room temperature for 4 weeks.
- Set up a similar experiment for each of the assayed culture media (R2A, TSB, LB, NB, 50% R2A, R2A-SC, J26-SE, and SCA-SE).
- After incubation, perform the sampling by taking 1 ml of culture broth from diffusion bioreactor and serially dilute the sample. For serial dilution, use freshly prepared normal saline.
- Spread 100 µl of serially diluted samples from each dilution (10-1-10-7) onto the agar plates for each respective medium. Perform each spreading in duplicates.
- Incubate aerobically at 25 °C for 4 weeks.
- After incubation, select the isolated colonies from each plate.
- Perform subculture repeatedly (at least twice) to obtain pure colonies.
- Identify pure colonies by 16S rRNA gene sequence analysis.
- PCR amplification and 16S rRNA gene sequencing
- Extract genomic DNA from the pure colonies using DNA isolation kit.
- Perform PCR amplification for 16S rRNA gene using the following primer set and conditions:
Primers (Frank et al., 2008)
Forward (27F), 5’-AGAGTTTGATCMTGGCTCAG-3’
Reverse (1492R), 5’-TACGGYTACCTTGTTACGACTT-3’
Reaction mixture
30 µl reaction mixture containing 20 ng of genomic DNA
PCR conditions
95 °C, 2 min
35 cycles: 95 °C, 1 min; 55 °C, 1 min; 72 °C, 1 min
72 °C, 10 min - Purify the PCR products using a Multiscreen PCR384 Filter Plate kit.
- Sequence the purified PCR products using a BigDye Terminator Cycle Sequencing Kit v.3.1 (Applied Biosystems) in a 3730XL DNA analyzer (Applied Biosystems).
Use the following primer set for sequencing reactions:
Forward (518F), 5′-CCAGCAGCCGCGGTAATACG-3′
Reverse (800R), 5′-TACCAGGGTATCTAATCC-3′ - Compile short sequence of 16S rRNA gene using SeqMan Pro (DNASTAR) software.
- Phylogenetic analysis
- Compare the sequenced 16S rRNA genes of all the isolates to find the closest phylogenetic neighbors using BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) and the EZBioCloud server (https://www.ezbiocloud.net) (Yoon et al., 2017).
- Retrieve the 16S rRNA gene sequences of closely related species from the GenBank.
- Construct the phylogenetic tree (Figure 3) in Mega 6.0.6 software using maximum-likelihood algorithm (Felsenstein, 1981; Tamura et al., 2013).
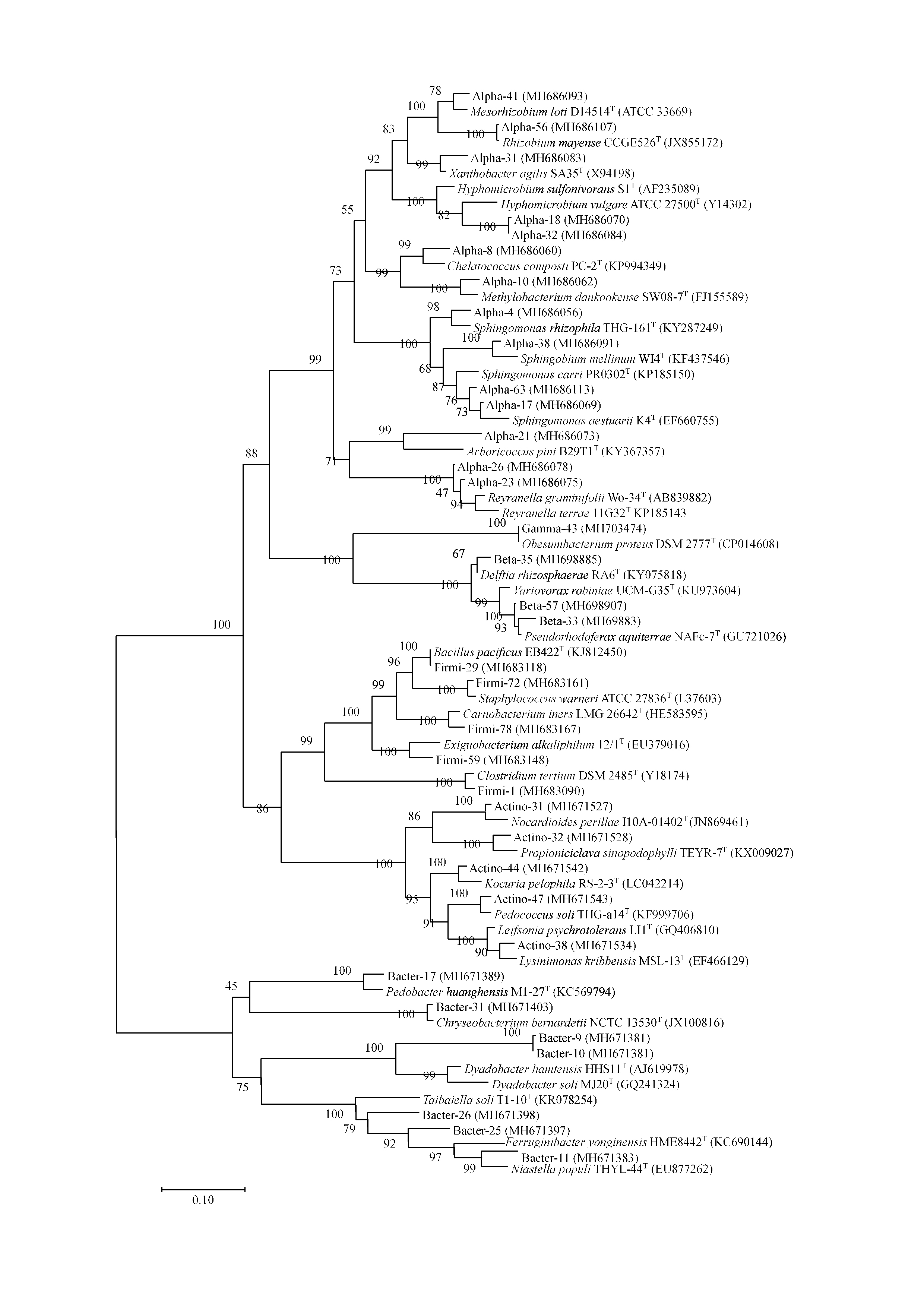
Figure 3. Phylogenetic tree. The phylogenetic tree was constructed using the maximum likelihood algorithm based on nearly complete 16S rRNA gene sequences. It shows the taxonomic affiliations of previously uncultured bacteria using the newly developed diffusion bioreactor with representatives of different established phylogenetic groups (Figure adapted from Chaudhary et al., 2019).
- Extraction of DNA from soil
- Extract DNA from soil samples using FastDNA SPIN kit (MP Biomedicals) following the manufacturer’s instructions.
- Assess the quality of DNA with 1.5% agarose gel electrophoresis.
- Determine the purity and concentration of extracted DNA using MaestroNano spectrophotometer (MaestroGen).
- PCR amplification and pyrosequencing
- Perform PCR amplification for V1-V3 regions of 16S rRNA gene using the following primer set and PCR conditions:
Primers (Hur et al., 2011; Okubo et al., 2012)
27F: 5′-CCTATCCCCTGTGTGCCTTGGCAGTC-TCAG-AC-GAGTTTGATCMTGGCTCAG-3′
518R: 5′-CCATCTCATCCCTGCGTGTCTCCGAC-TCAG-X-ACWTTACCGCGGCTGCTGG-3′; where ‘X’ represents unique barcode for each subject.
PCR conditions
30 °C, 5 min
30 cycles: 95 °C, 30 s; 72 °C, 30 s; 72 °C, 30 s
72 °C, 10 min - Assess the PCR amplicons by 1.5% agarose gel electrophoresis.
- Purify the PCR amplicons using QIAQuick PCR Purification Kit (Qiagen)
- Perform the pyrosequencing of the purified PCR products using Roche/454 GS FLX Titanium platform following the manufacturer’s instructions.
- Perform PCR amplification for V1-V3 regions of 16S rRNA gene using the following primer set and PCR conditions:
Data analysis
- Statistical analysis can be conducted using Microsoft Office Excel 2013 and OriginPro 8.5. The significant difference (p < 0.05) is estimated by two-way ANOVA analysis.
- The following threshold values can be used to infer novel taxa: species (99.0% ≥ x ≥96.5%), genus (96.5% > x ≥ 90%), family (90% > x ≥ 81.7%), order (81% > x ≥ 70%), and class (70% > x ≥ 65%), where ‘x’ represents the sequence similarity value (Yarza et al., 2008; Kim et al., 2014).
Notes
- For comparison of the effectiveness of diffusion bioreactors, set up traditional experiment of cultivation in similar conditions as illustrated in Figure 2.
- Sterilize all the components used to prepare diffusion bioreactors in 70% ethanol followed by drying under UV-chamber in a clean bench.
- This diffusion bioreactor relies on the type of soil sample to recover a diverse range of bacteria and other microbes. Selective uncultured bacteria can be cultivated in this bioreactor by using soil sampled from a specific area. For example, uncultured oil-contaminated bacteria can be recovered by using oil-contaminated soil.
Recipes
- Soil extract (SE)
- Mix 1 kg soil in 2 L sterile d/w
- Shake in an orbital shaker with 120 rpm shaking speed at room temperature overnight
- Stand for 2 h for settling
- Decant and collect supernatant
- Filter the supernatant through a 0.22 µm membrane filter
- Cultivation media (Table 1)
Table 1. Composition of media used for the cultivation (Chaudhary et al., 2019)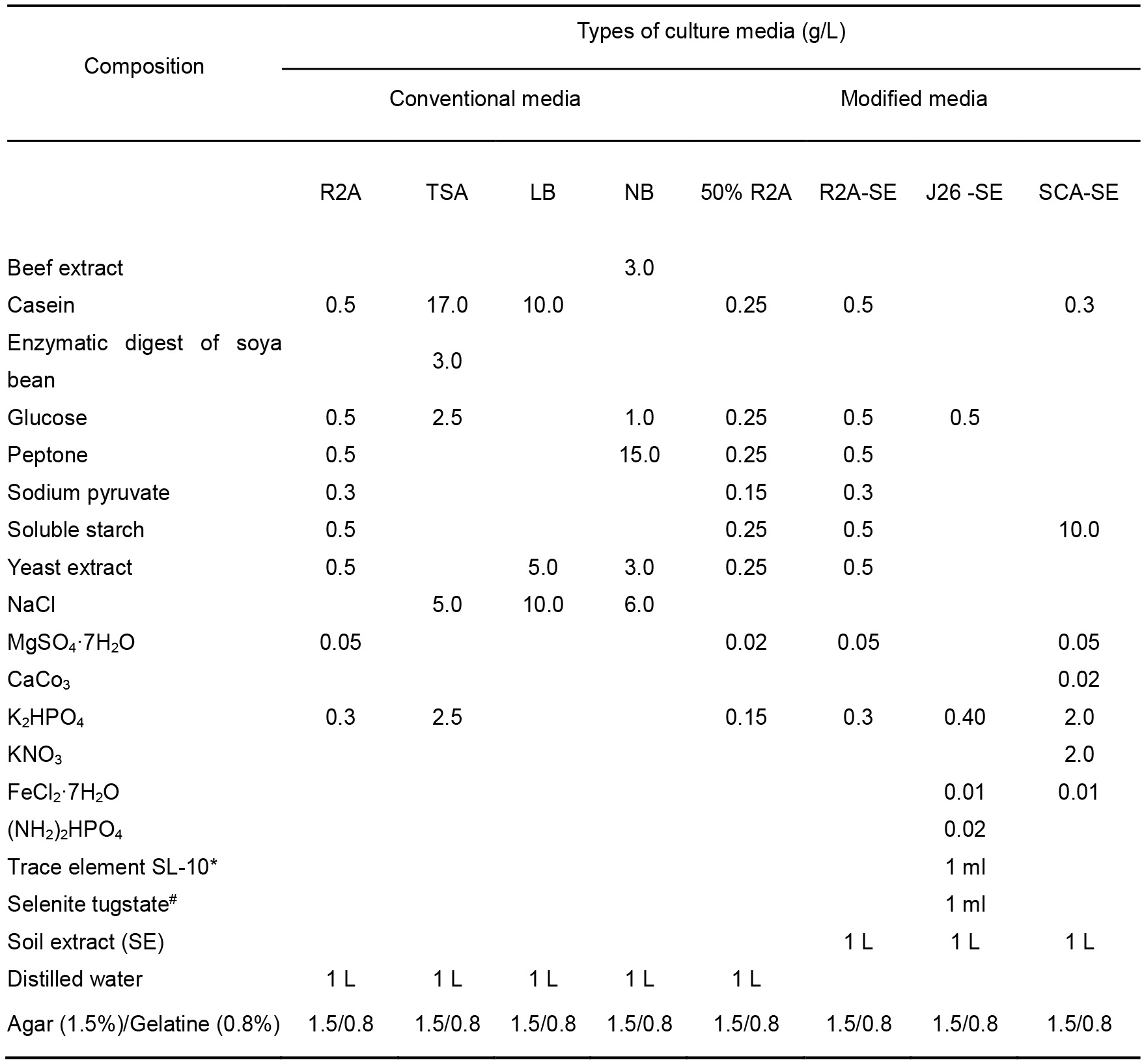
*see Recipe 3; #see Recipe 4.- Heat to boiling until completely dissolved.
- Adjust the pH to 7.2
- Sterilize at 121 °C for 15 min; pH= 7.2 ± 0.2 at 25 °C.
- Trace element SL-10
HCl, 10 ml
CoCl2·6H2O, 190 mg
CuCl2·2H2O, 2 mg
FeCl2·4H2O, 1.5 g
NaBO3, 6 mg
MnCl2·4H2O, 100 mg
Na2MoO4·2H2O, 36 mg
NiCl2·6H2O, 24 mg
ZnCl2, 70 mg
Distilled water, 990 ml
Filter through a 0.22 µm filter for sterilization - Selenite tugstate
NaOH, 0.5 g
Na2SeO3·5H2O, 3 mg
Na2WO4·2H2O, 4 mg
Distilled water, 1,000 ml
Filter through a 0.22 µm filter for sterilization
Acknowledgments
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF-2019R1F1A1058501). This protocol was adapted from Chaudhary et al., (2019) Development of a novel cultivation technique for uncultured soil bacteria. Scientific Reports, 9: 6666.
Competing interests
The authors declare there are no conflicts of interest.
References
- Bollmann, A., Lewis, K. and Epstein, S. S. (2007). Incubation of environmental samples in a diffusion chamber increases the diversity of recovered isolates. Appl Environ Microbiol 73(20): 6386-6390.
- Chaudhary, D. K., Khulan, A. and Kim, J. (2019). Development of a novel cultivation technique for uncultured soil bacteria. Sci Rep 9(1): 6666.
- Davis, K. E., Joseph, S. J. and Janssen, P. H. (2005). Effects of growth medium, inoculum size, and incubation time on culturability and isolation of soil bacteria. Appl Environ Microbiol 71(2): 826-834.
- Felsenstein, J. (1981). Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17(6): 368-376.
- Ferrari, B. C., Binnerup, S. J. and Gillings, M. (2005). Microcolony cultivation on a soil substrate membrane system selects for previously uncultured soil bacteria. Appl Environ Microbiol 71(12): 8714-8720.
- Frank, J. A., Reich, C. I., Sharma, S., Weisbaum, J.S., Wilson, B. A. and Olsen, G. J. (2008). Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA gene. Appl Environ Microbiol 74(8): 2461-2470.
- Gans, J., Wolinsky, M. and Dunbar, J. (2005). Computational improvements reveal great bacterial diversity and high metal toxicity in soil. Science 309(5739): 1387-1390.
- George, I. F., Hartmann, M., Liles, M. R. and Agathos, S.N. (2011). Recovery of as-yet-uncultured soil Acidobacteria on dilute solid media. Appl Environ Microbiol 77(22): 8184-8188.
- Hur, M., Kim, Y., Song H. R., Kim, J. M., Choi Y. I. and Yi, H. (2011). Effect of genetically modified poplars on soil microbial communities during the phytoremediation of waste mine tailings. Appl Environ Microbial 77(21): 7611-7619.
- Joseph, S. J., Hugenholtz, P., Sangwan, P., Osborne, C. A. and Janssen, P. H. (2003). Laboratory cultivation of widespread and previously uncultured soil bacteria. Appl Environ Microbiol 69(12): 7210-7215.
- Kaeberlein, T., Lewis, K. and Epstein, S. S. (2002). Isolating “uncultivable” microorganisms in pure culture in a simulated natural environment. Science 296(5570): 1127-1129.
- Kakumanu, M. L. and Williams, M. A. (2012). Soil diffusion system enriches the growth of diverse and previously uncultivated bacterial taxa. Soil Biol Biochem 76(2): 463-474.
- Kim. M., Oh, H. S., Park, S. C. and Chun, J. (2014). Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol 64(Pt 2): 346-351.
- Okubo, T., Ikeda, S., Yamashita, A., Terasawa, K. and Minamisawa, K. (2012). Pyrosequencing read length of 16S rRNA gene affects phylogenetic assignment of plant-associated bacteria. Microbes Environ 27(2): 204-208.
- Pham, V. H. and Kim, J. (2012). Cultivation of unculturable soil bacteria. Trends Biotechnol 30(9): 475-484.
- Sait, M., Hugenholtz, P. and Janssen, P. H. (2002). Cultivation of globally distributed soil bacteria from phylogenetic lineages previously only detected in cultivation-independent surveys. Environ Microbiol 4 (11): 654–666.
- Stewart, E. J. (2012). Growing unculturable bacteria. J Bacteriol 194(16): 4151-4160.
- Tamura, K., Stecher, G., Peterson, D., Filipski, A. and Kumar, S. (2013). MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30(12): 2725-2729.
- van Dorst, J. M., Hince, G., Snape, I. and Ferrari, B. C. (2016). Novel culturing techniques select for heterotrophs and hydrocarbon degraders in a Subantarctic soil. Sci Rep 6: 36724.
- Vartoukian, S. R., Palmer, R. M. and Wade, W. G. (2010). Strategies for culture of ‘unculturable’ bacteria. FEMS Microbiol Lett 309(1): 1-7.
- Yarza, P., Richter, M., Peplies, J., Euzeby, J., Amann, R., Schleifer, K. H., Ludwig, W., Glöckner F. O. and Rosselló-Móra, R. (2008). The All-Species Living Tree Project: A 16S rRNA-based phylogenetic tree of all sequenced type strains. Syst Appl Microbiol 31(4): 241-250.
- Yoon, S. H., Ha, S. M., Kwon, S., Lim, J., Kim, Y., Seo, H. and Chun, J. (2017). Introducing EzBioCloud: A taxonomically united database of 16S rRNA and whole genome assemblies. Int J Syst Evol Microbiol 67(5): 1613-1617.
Article Information
Copyright
© 2019 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Chaudhary, D. K. and Kim, J. (2019). Experimental Setup for a Diffusion Bioreactor to Isolate Unculturable Soil Bacteria. Bio-protocol 9(19): e3388. DOI: 10.21769/BioProtoc.3388.
Category
Microbiology > Microbial cell biology > Cell isolation and culture
Environmental science > Bacterium > Isolation
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











