- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
3D Organoid Formation from the Murine Salivary Gland Cell Line SIMS
Published: Vol 9, Iss 19, Oct 5, 2019 DOI: 10.21769/BioProtoc.3386 Views: 6245
Reviewed by: Giusy TornilloJuan Mauricio GarréAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

A Simplified 3D-Plasma Culture Method for Generating Minimally Manipulated Autologous Equine Muscle-Derived Progenitor Cells
Hélène Graide [...] Didier Serteyn
Dec 5, 2025 1251 Views

Optimization of Adipogenic Differentiation Protocol for Murine and Human Cell Culture Models
Junwan Fan [...] Wenyan He
Jan 20, 2026 217 Views

Simple and Rapid Model to Generate Differentiated Endometrial Floating Organoids
Adriana Bajetto [...] Tullio Florio
Feb 5, 2026 97 Views
Abstract
Salivary glands consist of multiple phenotypically and functionally unique cell populations, such as the acinar, ductal, and myoepithelial cells that help produce, modify, and secrete saliva (Lombaert et al., 2011). Identification of mechanisms and factors that regulate these populations has been of key interest, as salivary gland-related diseases have detrimental effects on these cell populations. A variety of approaches have been used to understand the roles different signaling mechanisms and transcription factors play in regulating salivary gland development and homeostasis. Differentiation assays have been performed with primary salivary cells in the past (Maimets et al., 2016), however this approach may sometimes be limiting due to tissue availability, labor intensity of processing the tissue samples, and/or inability to long-term passage the cells. Here we describe in detail a 3D differentiation assay to analyze the differentiation potential of a salivary gland cell line, SIMS, which was immortalized from an adult mouse submandibular salivary gland (Laoide et al., 1996). SIMS cells express cytokeratin 7 and 19, which is characteristic for a ductal cell type. Although adult acinar and myoepithelial cells were found in vivo to preserve their own cell population through self-duplication (Aure et al., 2015; Song et al. 2018), in some cases duct cells can differentiate into acinar cells in vivo, such as after radiation injury (Lombaert et al., 2008; Weng et al., 2018). Thus, utilization of SIMS cells allows us to target and analyze the self-renewal and differentiation effects of ductal cells under specific in vitro controlled conditions.
Keywords: Salivary glandBackground
In vitro differentiation assays are a remarkable tool that enables us to analyze the potency of cells without relying on in vivo models. Cells in these assays can be manipulated for gene expression to, for instance, analyze their differentiation ability. While previous protocols have described the differentiation potential of primary salivary gland cells in vitro, our protocol is optimized for the use of the SIMS cell line (Laoide et al., 1996; Maimets et al., 2016). SIMS cells express cytokeratin 7 and 19, which is characteristic for a ductal cell type. Although adult acinar and myoepithelial cells were found in vivo to preserve their own cell population through self-duplication (Aure et al., 2015; Song et al. 2018), in some cases duct cells can differentiate into acinar cells in vivo, such as after radiation injury (Lombaert et al., 2008; Weng et al., 2018). Thus, utilization of SIMS cells allows us to target and analyze the self-renewal and differentiation effects of ductal cells under specific in vitro controlled conditions. Our recent work shows that SIMS cells have the ability to differentiate into unique populations of acinar, myoepithelial, and duct cells in 3D differentiation conditions, that normally produce, modify and secrete saliva in vivo (Lombaert et al., 2011; Athwal et al., 2019). This protocol describes the workflow necessary for the generation and analysis of 3D differentiated SIMS cells. SIMS cells are cultured in a 3D matrigel/collagen matrix over a period of 7-9 days. Matrigel is a solubilized basement membrane preparation extracted from the Engelbreth-Holm-Swarm (EHS) mouse sarcoma, which is rich in several growth factors and extracellular matrix proteins, such as laminin-1, collagen IV, and heparin sulfate proteoglycans (Patel et al., 2016). Matrigel has been shown to induce differentiation of stem/progenitor cells and outgrowth of already differentiated cell types by causing polarization of cells embedded in or on top of it. For SIMS grown in this 3D matrix, organoid formation post single cell culture is apparent around day 5 of culture. These organoids are also stained either via cryosectioning or in 3D to analyze for differentiation markers and cell populations. Alterations, such as cell transfections, transductions and/or media changes, to this condition can easily be made to address specific research questions (Athwal et al., 2019). The media components were adjusted using literature from both existing salivary and mammary gland differentiation protocols, which deemed necessary to induce differentiation of the SIMS cells in vitro. Here we show the robust use of this cell line to address specific research questions without having to rely on primary salivary gland cells.
Materials and Reagents
- 500 ml filter units with 0.22 μm filter (Fisher, catalog number: 974102)
- 24-well tissue culture plates (Thermo Fisher, catalog number: 142475)
- T75 tissue culture flasks (Thermo Fisher, catalog number: 156499)
- 2 ml pipettes (Fisher, catalog number: 13-678-25C)
- 10 cm dishes (Thermo Fisher, catalog number: 150464)
- 15 ml Falcon tubes (Corning, catalog number: 35296)
- 1 ml pipettes (Fisher, catalog number: 13-678-25B)
- Forceps (Fisher Scientific, catalog number: XX6200006P)
- Razor blade (Fisher Scientific, catalog number: 12-640)
- Glass slides (VWR, catalog number: 89500-466)
- SIMS cells (available per request)
- Sterile DMEM medium + 4.5g/L high glucose + L-glutamine without sodium pyruvate (Invitrogen, catalog number: 11965-092)
- Sterile 1x PBS-Ca+2-Mg+2 free (Thermo Fisher, catalog number: 110010031) or 1x HBSS-Ca+2-Mg+2 free (Thermo Fisher, catalog number: 14175079)
- Penicillin-Streptomycin (Thermo Fisher, catalog number: 15140122)
- Sterile USDA-approved FBS (Sigma-Aldrich, catalog number: F2442-500ML)
- 0.25% Trypsin-EDTA (Sigma-Aldrich, catalog number: T4049-100ML)
- Epidermal growth factor (EGF) (Sigma-Aldrich)
- Glutamax (Gibco/ThermoFisher, catalog number: 35050061)
- Fibroblast growth factor-2 (FGF2) (R&D, catalog number: 3139FB025)
- N2 (Thermo Fisher, catalog number: 17502048)
- Dexamethasone (VWR, catalog number: 80056-298)
- Triiodothyronine (Sigma-Aldrich, catalog number: 709719)
- Retinoic Acid (Thermo Fisher, catalog number: AA4454077)
- Hydrocortisone (Sigma-Aldrich, catalog number: H0888)
- Cholera toxin (Sigma-Aldrich, catalog number: C8052)
- Calcium, 99%, Granular (Fisher, catalog number: AC201180050)
- Growth factor reduced Matrigel (BD Biosciences, catalog number: 354230)
- Collagen Type I, Rat Tail (Millipore, catalog number: 2790888)
- 4% PFA (VWR, catalog number: AAJ61899-AK)
- Trypan blue 0.4% (Thermo Fisher, catalog number: 15250061)
- O.C.T. Compound (Fisher scientific, catalog number: 23-730-571)
- SIMS subculture media (see Recipes)
- Differentiation media (see Recipes)
Equipment
- Water bath (Thermo Scientific, catalog number: TSGP02)
- Vacuum pump aspirator (standard)
- CO2 incubator (standard)
- Cryotome (standard)
- Brightfield microscope (standard)
- Confocal laser scanning microscope (standard)
Software
- ImageJ software (http://imagej.net, available online for download)
Procedure
- SIMS subculture
- Pre-warm cell medium, 1x PBS-Ca+2-Mg+2 free or 1x HBSS- Ca+2-Mg+2 free and 0.25% trypsin-EDTA in a water bath at 37 °C.
- Aspirate off old medium with vacuum aspirator or use 10 ml pipettes to manually aspirate from the T75 cell culture flask.
- Wash twice with 10 ml of sterile 1x PBS/HBSS-Ca+2-Mg+, aspirate off completely-either manually with a 2 ml pipette or with an aspirator and sterile fresh 2 ml glass pipette.
- Add 3 ml of pre-warmed 0.25% Trypsin-EDTA per T75 flask.
- Incubate plate with cells at 37 °C for 5-7 min. Tap and gently shake the plate to dislodge cells.
- Neutralize trypsin by adding fresh ~3-5 ml culture medium using a 10 ml pipette. Pipette gently up and down several times to re-suspend cells and transfer to a 15 ml Falcon tube.
- Centrifuge at 100 x g for 3 min, wash 2 times with 1x PBS-Ca+2-Mg+2 free, and take up the cells in desired amount of culture medium.
- Plate up to 1 million cells to get 95% confluency in 4-5 days in a T75 tissue culture flask.
Note: Cell viability (~90-95%) is determined by trypan blue cell count.
- Organoid differentiation culture
- Resuspend SIMS cells in SIMS medium at 0.4 x 106 cells/ml.
- Thaw out growth factor reduced matrigel matrix on ice for 10 min before culturing cells. Both collagen and matrigel need to remain on ice until they are resuspended with cells for plating.
- Add 50 μl of cell solution (10,000 cells) to 100 µl of 60:40 ratio of Type I rat tail collagen to growth factor reduced Matrigel on ice. Mix gently while slowly pipetting up and down using a 200 μl pipette.
Notes:- Aliquot collagen first then Matrigel. Do not mix with pipette until you add cells to avoid bubbles.
- Avoid fast aspiration or suspension of aliquoted matrix mix to avoid bubble formation.
- Deposit cell/collagen/matrigel mix in the center of 24-well tissue culture plate on ice and incubate for 20 min at 37 °C.
- Add up to 1 ml differentiation medium.
- Change medium every 2-3 days.
- Cultures are maintained up to 14 days as the cells reach maximum confluency within the matrix, and the matrix starts breaking down.
- Organoid formation can be seen starting Day 5 of culture (Figure 1).
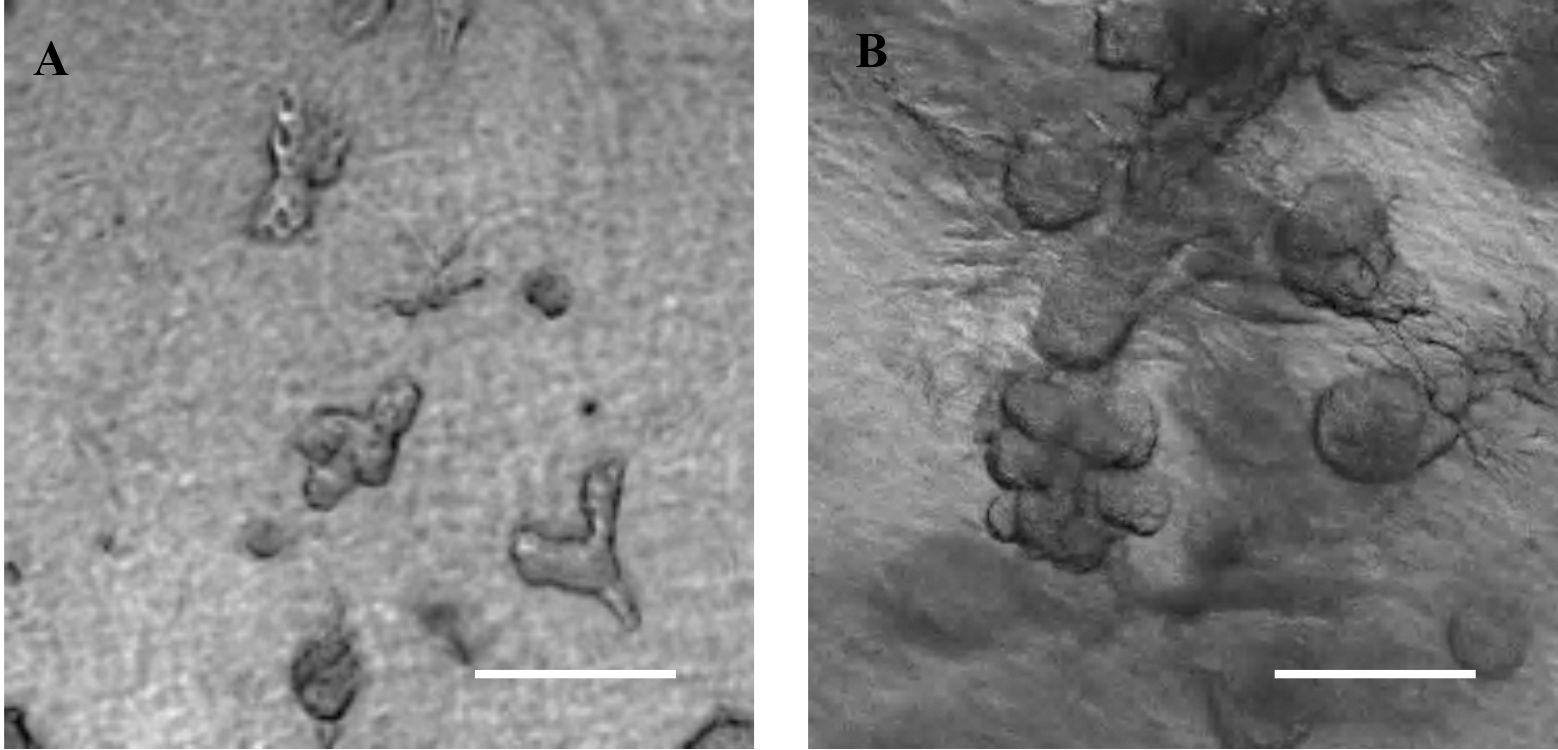
Figure 1. Collagen-matrigel embedded SIMS cells expand and form organoids by Day 5 (A) and 7 (B). Scale bars, 200 μm (4x magnification).
- Organoid fixation
- Wash matrix with ice cold 1x PBS-Ca+2-Mg+2 free gently up to 3 times.
- Cells can be fixed at any timepoint using 2% PFA in 1x PBS-Ca+2-Mg+2 free for 20 min on ice or at 4 °C.
- Rinse the matrix with ice cold 1x PBS-Ca+2-Mg+2 free 3 times.
- Incubate matrix in 1x PBS-Ca+2-Mg+2 free for 10 min. Repeat this 3 times for a total of 30 minutes.
- Using forceps, gently remove the matrix from the tissue culture plate.
Note: You can cut the matrix into 2-3 pieces using a fine blade if you want multiple cryo blocks. - Embed with OCT (a full description of embedding can be found in Campbell et al., 2011).
Note: Matrix will likely curl up in OCT if it is poured too quickly. Gently pour, and fix any curling using your forceps. - Optimal cutting thickness is between 12 and 16 μm.
- Sections can be post fixed with 4% PFA, ice cold acetone, or acetone/methanol for staining optimization.
- 3D matrix staining
To conserve the 3D morphology, staining is performed on 1 mm3 pieces of the organoids embedded in matrix.- Post fixation, small pieces of matrix including organoids are cut using a sharp scalpel blade and pointed tweezers. Treat each chunk separately in a plastic dish or 24-well plate.
- Matrix can be treated again with 2-4% PFA, acetone/methanol, or methanol treatments if necessary.
- Matrix is treated like tissue slides for staining. An example is described in Athwal et al., 2019.
- Mount each sample on glass slides with 1-2 spacers depending on the size of the matrix.
- Capture images using a confocal microscope (Figure 2).
- This staining can also be performed on cryosections of OCT embedded organoid matrix (Athwal et al., 2019). A complete description of the primary and secondary antibodies used in Figure 2 can be found in above-mentioned manuscript.
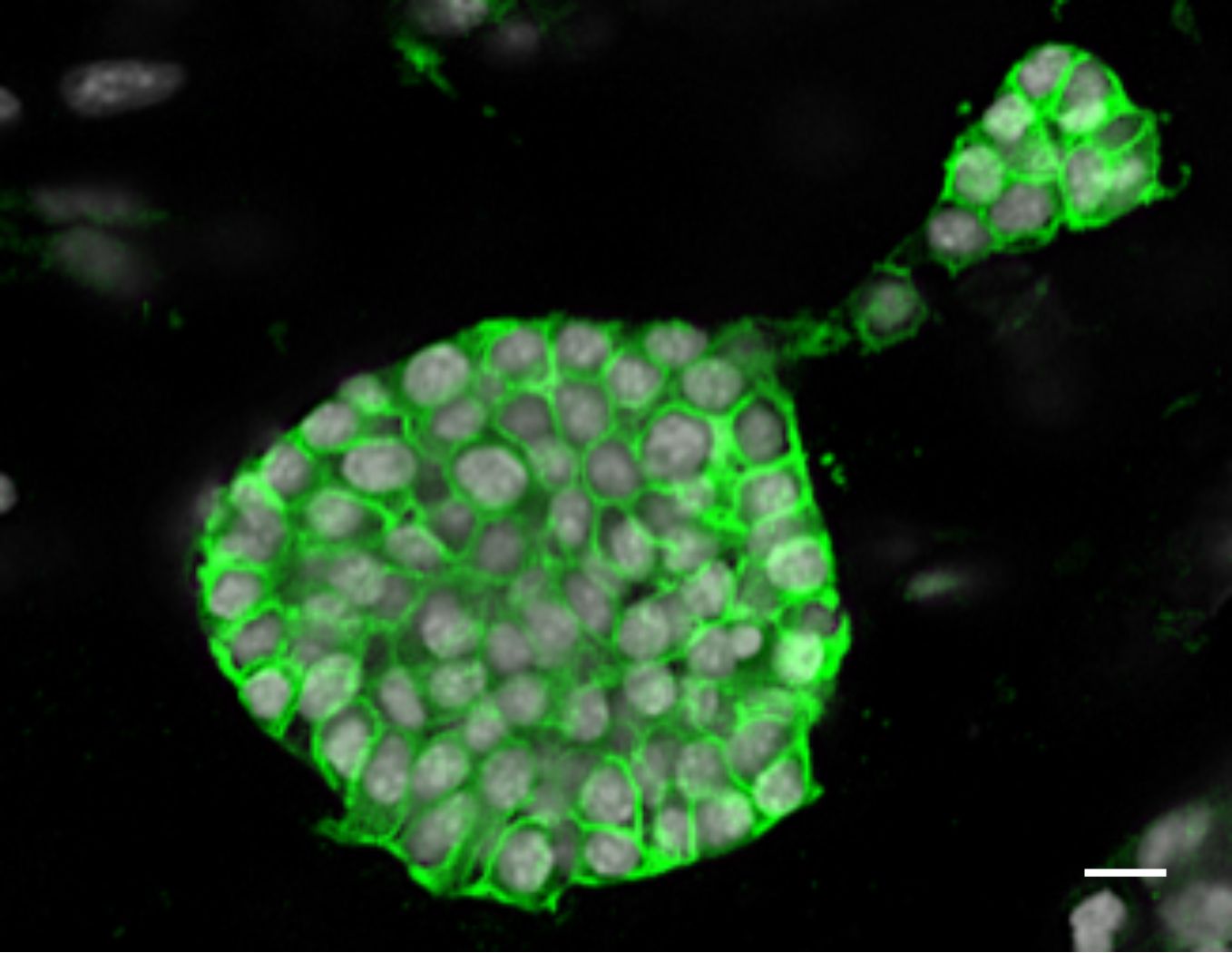
Figure 2. 1 mm3 collagen/matrigel matrix with 3D grown SIMS organoids were stained with E-cadherin post-fixation with 4% PFA. Scale bar, 10 μm.
Data analysis
Brightfield images are taken starting Day 3 until the end of the experiment. Organoid diameter can be quantified using ImageJ. Each experiment is repeated 3 times with n = 5 images per day for quantification. Details on statistical analysis can be found in our previous manuscript (Athwal et al., 2019).
Notes
- Cell death is noticed by Day 14 as cells become confluent within the matrix.
- Serial passage of cells dissociated from organoids has not been performed with SIMS cell organoids.
- Increase the volume of collagen/matrigel if cell number is increased.
- Transduced or transfected SIMS cells are selected prior to the 3D culture assay.
Recipes
- SIMS subculture medium (Table 1)
Table 1. SIMS subculture medium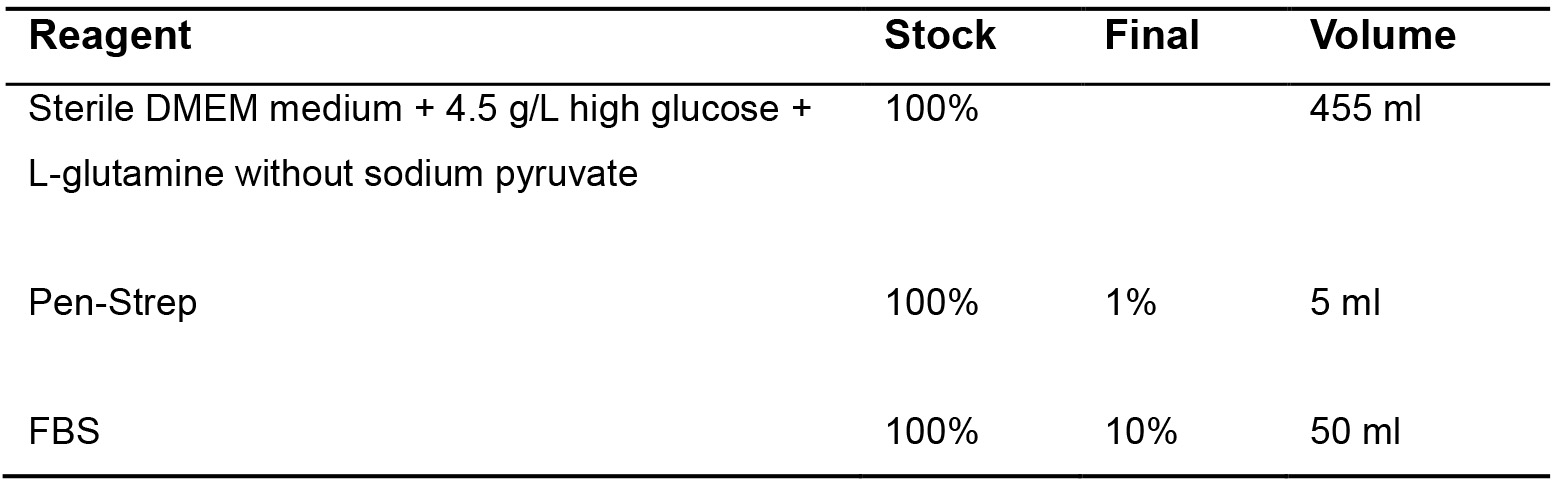
- Differentiation medium
Table 2. SIMS differentiation medium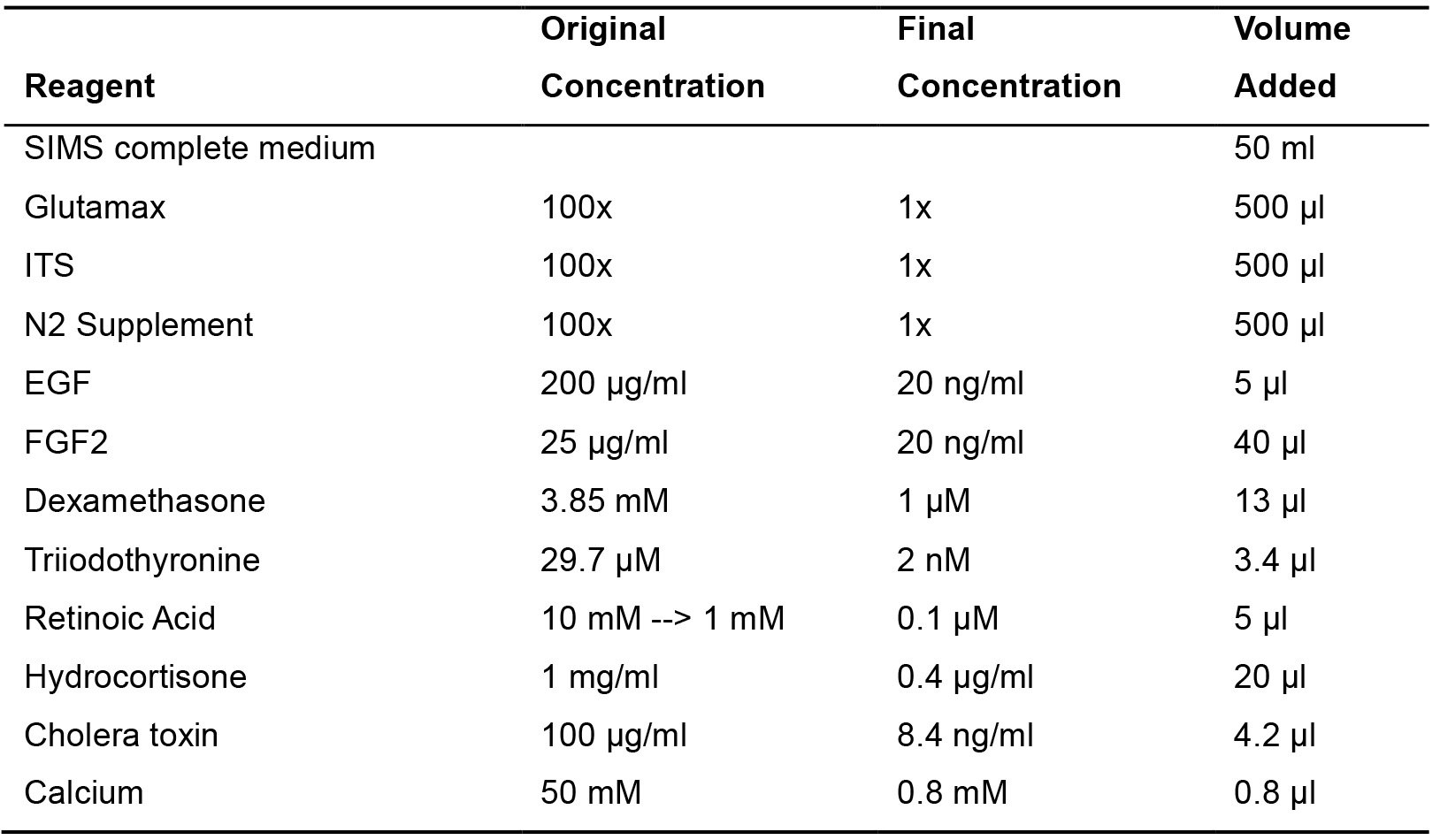
Notes:- Make 100 μl aliquots of growth factor reduced matrigel and store at -20 °C.
- Make 100 μl aliquots of rat tail collagen and store at 4 °C.
Acknowledgments
This work was supported by the National Institutes of Health (NIH)/National Institute of Dental and Craniofacial Research (NIDCR) grants DE027034 and DE022557. This protocol was adapted from Maimets et al., 2016. Timeline of organoid formation using SIMS cells and changes in media recipe are described in our protocol.
Competing interests
There are no conflicts of interest.
References
- Athwal, H. K., Murphy, G., 3rd, Tibbs, E., Cornett, A., Hill, E., Yeoh, K., Berenstein, E., Hoffman, M. P. and Lombaert, I. M. A. (2019). Sox10 regulates plasticity of epithelial progenitors toward secretory units of exocrine glands. Stem Cell Reports 12(2): 366-380.
- Aure, M. H., Konieczny, S. F. and Ovitt, C. E. (2015). Salivary gland homeostasis is maintained through acinar cell self-duplication. Dev Cell 33(2): 231-237.
- Campbell, J. J., Davidenko, N., Caffarel, M. M., Cameron, R. E. and Watson, C. J. (2011). A multifunctional 3D co-culture system for studies of mammary tissue morphogenesis and stem cell biology. PLoS One 6(9): e25661.
- Laoide, B. M., Courty, Y., Gastinne, I., Thibaut, C., Kellermann, O. and Rougeon, F. (1996). Immortalised mouse submandibular epithelial cell lines retain polarised structural and functional properties. J Cell Sci 109 (Pt 12): 2789-2800.
- Lombaert, I. M., Brunsting, J. F., Wierenga, P. K., Faber, H., Stokman, M. A., Kok, T., Visser, W. H., Kampinga, H. H., de Haan, G. and Coppes, R. P. (2008). Rescue of salivary gland function after stem cell transplantation in irradiated glands. PLoS One 3(4): e2063.
- Lombaert, I. M., Knox, S. M. and Hoffman, M. P. (2011). Salivary gland progenitor cell biology provides a rationale for therapeutic salivary gland regeneration. Oral Dis 17(5): 445-449.
- Maimets, M., Rocchi, C., Bron, R., Pringle, S., Kuipers, J., Giepmans, B. N., Vries, R. G., Clevers, H., de Haan, G., van Os, R. and Coppes, R. P. (2016). Long-term in vitro expansion of salivary gland stem cells driven by Wnt signals. Stem Cell Reports 6(1): 150-162.
- Patel, R. and Alahmad, A. J. (2016). Growth-factor reduced Matrigel source influences stem cell derived brain microvascular endothelial cell barrier properties. Fluids Barriers CNS 13: 6.
- Song, E. C., Min, S., Oyelakin, A., Smalley, K., Bard, J. E., Liao, L., Xu, J. and Romano, R. A. (2018). Genetic and scRNA-seq analysis reveals distinct cell populations that contribute to salivary gland development and maintenance. Sci Rep 8(1): 14043.
- Weng, P. L., Aure, M. H., Maruyama, T. and Ovitt, C. E. (2018). Limited regeneration of adult salivary glands after severe injury involves cellular plasticity. Cell Rep 24(6): 1464-1470 e3.
Article Information
Copyright
© 2019 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Athwal, H. K. and Lombaert, I. M. A. (2019). 3D Organoid Formation from the Murine Salivary Gland Cell Line SIMS. Bio-protocol 9(19): e3386. DOI: 10.21769/BioProtoc.3386.
Category
Developmental Biology > Morphogenesis > Organogenesis
Stem Cell > Adult stem cell > Maintenance and differentiation
Cell Biology > Cell isolation and culture > 3D cell culture
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link