- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Using indCAPS to Detect CRISPR/Cas9 Induced Mutations
Published: Vol 9, Iss 18, Sep 20, 2019 DOI: 10.21769/BioProtoc.3374 Views: 4389
Reviewed by: Rainer MelzerMarcelo Santos da SilvaAyush Ranawade

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Reference-free Association Mapping from Sequencing Reads Using k-mers
Zakaria Mehrab [...] Atif Rahman
Nov 5, 2020 5466 Views

Conditional Human BRD4 Knock-In Transgenic Mouse Genotyping and Protein Isoform Detection
Michael Paul Lewis [...] Cheng-Ming Chiang
Apr 5, 2022 3618 Views

CAPS-Based SNP Genotyping for Nitrogen-Response Phenotypes in Maize Hybrids
Jannis Jacobs [...] Peter K. Lundquist
Dec 20, 2025 543 Views
Abstract
Cleaved amplified polymorphic sequences (CAPS) assays are useful tools for detecting small mutations such as single nucleotide polymorphisms (SNPs) or insertion/deletions (indels) present in an amplified DNA fragment. A mutation that disrupts or creates a restriction site will prevent cleavage by a restriction enzyme, allowing discrimination of wild-type and mutant alleles. In cases where no convenient restriction site is present, a derived Cleaved Amplified Polymorphic Sequence (dCAPS) assay can be used, where mismatches in the primer are used to create a diagnostic restriction site. No special design constraints are present for a CAPS assay, but cases where CAPS assays can be used are infrequent. A dCAPS assay can be burdensome to design by hand, but it is more broadly applicable. This protocol will describe the use of the indCAPS tool for the design of CAPS and dCAPS primers. The indCAPS tool was designed to be compatible with indel alleles, which prior tools struggled with but have increased importance since the rise of CRISPR/Cas9 mutagenesis methods.
Keywords: CRISPRBackground
Detecting mutations in biological samples is a common task in biological research. This is often performed using polymerase chain reaction (PCR) assays, where a sample’s genotype at one locus is determined by examining the size or sequence of amplified DNA. This protocol is a practical guide to using the indCAPS tool (Hodgens et al., 2017) available at http://indcaps.kieber.cloudapps.unc.edu/ to generate primers for PCR-based genotyping tasks. The indCAPS tool relies on a pair of techniques known as Cleaved Amplified Polymorphic Sequences (CAPS) and derived CAPS (dCAPS) (Neff et al., 1998), in which the genotype of a biological sample is determined by amplifying a region of DNA and assaying a diagnostic restriction site with a restriction enzyme. In a CAPS assay, the mutation of interest disrupts a known restriction site or creates a restriction site where one did not exist. In a dCAPS assay, a diagnostic site is created by introducing an intentional mismatch to one of the primers used to amplify the DNA, creating a restriction site in one genotype but not the other. Tools for generating primers for CAPS and dCAPS assays have been developed (Neff et al., 1998 and 2002) but are not compatible with insertion/deletion (indel) alleles such as those commonly acquired through CRISPR/Cas9 mutagenesis (Hodgens et al., 2017). With the recent rise of indel alleles in the literature due to CRISPR/Cas9 mutagenesis, a tool capable of generating dCAPS primers for indel alleles would make genotyping tasks easier. The indCAPS tool was developed to provide a tool for generating CAPS and dCAPS primers compatible with indels (Hodgens et al., 2017) and is intended to be easy to use and interpret for biologists designing genotyping assays. This protocol will describe the use of the indCAPS website for generating primers for genotyping a known polymorphism as well as detecting a novel mutation in a CRISPR/Cas9 mutagenesis experiment.
Equipment
- Computer
A computer capable of accessing the web. Desktops or laptops may be preferable to aid copying and pasting long DNA sequences. Tested on Windows and Mac computers. Linux computers should be compatible as well.
Software
- indCAPS tool (http://indcaps.kieber.cloudapps.unc.edu/)
Procedure
For screening for CRISPR/Cas9 alleles:
The genomic sequence of the targeted edit site, with 30-40 bases on each side of the predicted cut site. The CRISPR/Cas9 target sequence does not need to be in the same orientation as the DNA sequence, but it must be arranged such that the protospacer-adjacent motif (PAM) would appear to the right of the provided sequence. For example, the CRISPR/Cas9 target sequence GGTTGAGATCAAGATAGACA would be immediately followed by an NGG PAM to yield GGTTGAGATCAAGATAGACANGG. Only the target sequence (underlined) should be provided for these tools. An example of the sequences required can be seen in Figure 1.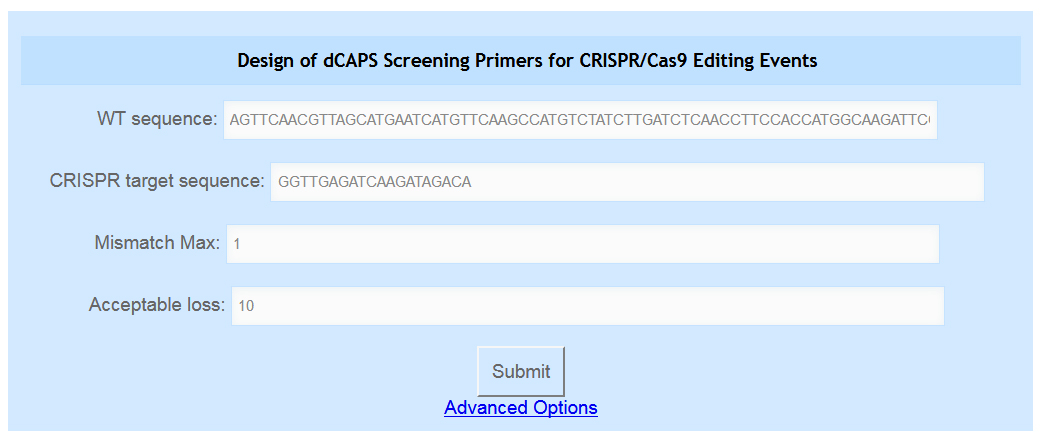
Figure 1. The Screening Primers tool. The user can enter their wild-type sequence in the first text box and their CRISPR target sequence in the second text box. Their desired maximum number of mismatches and the acceptable loss parameter are entered in the third and fourth boxes. Advanced options are accessed by clicking the blue “Advanced Options” link.
- Primer design for detection of unknown CRISPR/Cas9-derived alleles
- Navigate to the indCAPS main page and examine the second blue box. The box is labeled “Design of dCAPS Screening Primers for CRISPR/Cas9 Editing Events”. The fields are populated with example sequences.
- Copy the wild type genomic sequence and CRISPR target into the fields labeled “WT Sequence” and “CRISPR target sequence”. The copied sequence should not include any FASTA identifier lines (starting with a > character).
- Adjust parameters as necessary:
- Mismatch Max: The mismatch max parameter affects how many mismatching bases are allowed in a reported primer. If this value is exceeded, the primer is rejected. The mismatch value can be zero or any positive integer.
- Acceptable loss: This is the proportion of editing events the user is willing to miss with this assay. The indCAPS tool simulates the CRISPR mutagenesis process to generate a set of predicted possible mutants. In some cases, one of the edited fragments might not be diagnostic. For example, DraI cuts bluntly at TTT^AAA sites. Consider the case of a DraI site in the following sequence, where the insertion or deletion occurs at the | symbol: GCTTTAA|AAC. A single base deletion or an insertion of a single A will not disrupt the restriction site, while an insertion of a G, C, or T will create a diagnostic site. By default, if the proportion of all predicted mutations that create undiagnostic sites is above 10%, the assay is rejected. If the proportion is less than 10%, the assay can proceed. If the user does not wish to tolerate missed mutations, setting this parameter to 0 will ensure all reported assays find all predicted mutations.
- Amplicon Length: This adjusts how long your amplified fragment is expected to be. The indCAPS tool examines the sequence downstream of the primer it generates to ensure that no exact matches of the restriction site are present. For example, if you supplied 150 bases on each side of the mutation of interest, then with the default value of 90 bases, only the 90 bases downstream of the primer will be examined for restriction sites.
- Primer Type: This binary choice determines whether primers are generated to a specific length or to a specific melting temperature. Melting temperature is the default. If length is chosen, the value for desired melting temperature (item A.3.f. following this entry) is ignored when generating primers. Similarly, if the melting temperature is chosen, then the desired length value (item A.3.e) is ignored when generating primers.
- Primer Length: Desired length of the primer if the Primer Type is set to Length.
- Melting Temperature: Desired melting temperature if the Primer Type is set to TM.
- Allow 3’ G/T Mismatches: By default, indCAPS ignores any primers with terminal 3’ mismatches. Some literature reports that certain 3’ mismatches like G/T are compatible with PCR. To allow those mismatches in primer design, set this parameter to Yes.
- Salt Concentration: This value is used to calculate the melting temperature of the primers. A default value of 50 mM is used.
- Primer Concentration: This value is used to calculate the melting temperature of the primers. A default value of 50 mM is used.
- Press “Submit”.
- Examine the output:
- If any warnings are generated, they will be present in a bulleted list at the top of the output page. Warnings may indicate the presence of unexpected characters in the input or insufficiently long DNA sequences.
- Individual reports are separated by a series of “=” signs.
- Some reports may say “Sequences are reversed.” This means the sequences are displayed as the reverse-complement of what the user submitted and the primer is a reverse primer relative to the original sequence.
- The Enzyme name is listed.
- The next two lines indicate whether any matches to the current restriction enzyme motif were detected. The values in brackets are base positions, counting from the left of the displayed sequence. These values are useful if the user wants to identify the position of restriction motifs in the sequence by hand. A “Possible cut site” is a potentially diagnostic site that could be used in a CAPS or dCAPS assay. A “Problem cut site” is an exact match to the restriction motif present upstream of the diagnostic site. These sites are problems because they will be cleaved in both wild-type and mutant alleles, shortening the length of the fragment used to genotype the sequences. Problem sites will be disrupted by mismatches in the primer so that the ability to resolve diagnostic sequences is not impaired.
- The next two lines indicate the user-supplied sequences.
- The two following lines show the position of the restriction motif aligned to the two sequences as well as the aligned primer generated by the tool. Lowercase letters indicate mismatches introduced to the primer.
- If a problem site is found, a series of “.” Symbols will be aligned with the two sequences indicating its position.
- The last line indicates which sequence is cut and the estimated melting temperature for the primer.
- A sample output is shown in Figure 2.
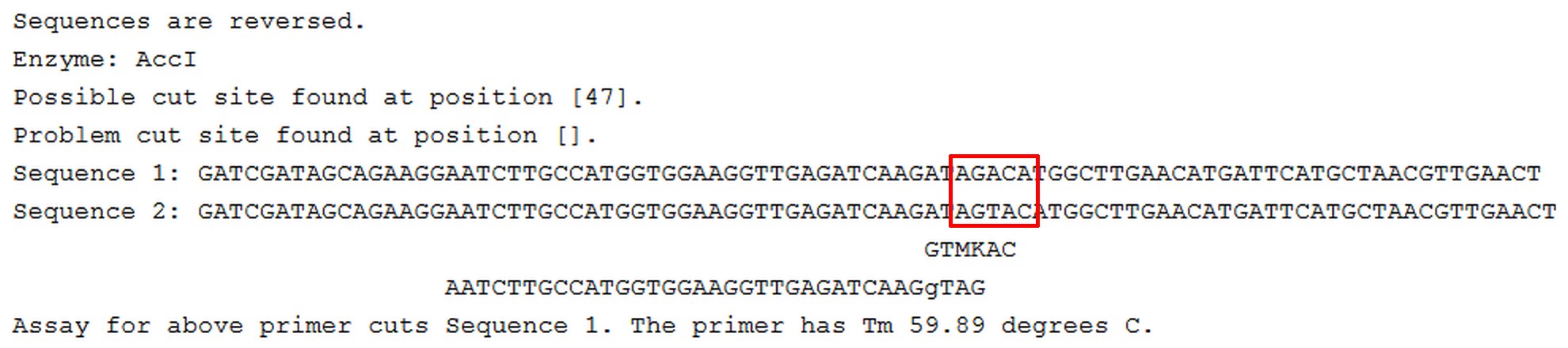
Figure 2. Sample indCAPS output. This is a reverse complement to the user-supplied sequence, showing a diagnostic primer intended to be used with an AccI restriction enzyme digestion. The supplied sequences are aligned with the identified restriction site and the suggested primer (including any mismatches). The mutation is indicated with a red box. - Primer selection criteria:
All technically possible assays are reported without regard for enzyme price or enzyme reliability and it can be difficult to predict how much a mismatch in the primer will affect the reaction. A CAPS assay is recommended over a dCAPS assay where possible, as size differences in the digested fragments are often easier to resolve.
Several recommendations:- If using a dCAPS assay, choose a primer with mismatches as far from the 3’ end as possible.
- If many dCAPS assays are to be performed at the same time, it is useful to save all results for each assay. By comparing the lists, you may be able to choose primers that use the same, inexpensive enzyme for multiple assays rather than a different, expensive enzyme for each assay.
- If using a CAPS assay, the user can also use primers equidistant from the diagnostic site rather than the suggested primer which abuts the diagnostic site.
Restriction motifs with a series of degenerate bases in the center such as BslI (CCNNNNNNNGG) are especially useful. If the CRISPR/Cas9 cleavage site is in the middle of the degenerate bases, the resulting indel often changes the number of bases between the CC/GG flanking sequences resulting in failure of the digest reaction.
For genotyping of known alleles:
Genomic sequence containing the mutant and wild-type alleles. The sequence should be centered on the mutation of interest with 30-40 bases flanking on each side, for a total length of 60-80 bases. The two sequences should be identical at the start and end, with the only difference being the mutation of interest in the middle.
Note: The two sequences can be shorter or longer as desired. If the sequences are too short, the primers generated by indCAPS may not have acceptable melting temperatures, and the user will have to manually extend the primers.
- Primer design for genotyping of known alleles
- Navigate to the indCAPS main page and examine the first blue box, at the top of the page and shown in Figure 3. The box is labeled “Design of dCAPS Primers for Known Alleles”. The fields are populated with example sequences.
- Copy your sequences into the two fields labeled “First Sequence” and “Second Sequence”.
- Adjust parameters as necessary. The parameters are similar to those described by the CRISPR/Cas9 screening tool with the exception that no Acceptable Loss parameter is present.
- Press “Submit”.
- The output and selection criteria are the same as those described for the CRISPR/Cas9 tool.
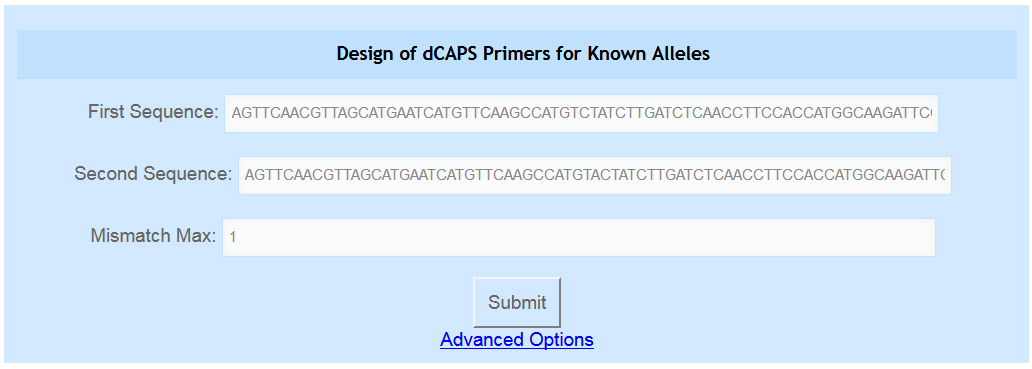
Figure 3. The Known Alleles tool. The user can enter their two sequences in the first two text boxes and indicate their desired maximum number of mismatches in the third box. Advanced options are accessed by clicking the blue “Advanced Options” link.
- Primer design for screening for isogenic mutagenic events
- This tool, shown in Figure 4, is intended for a rarer use case where CRISPR/Cas9 mutagenesis is used to isolate a mutant isogenic to a previously known mutation. In practice, its use is similar to the Known Alleles tool, where CAPS/dCAPS primers are designed that distinguish two sequences at a diagnostic site. An additional step is taken where the primers are evaluated to determine whether only desired mutations are detected by the assay.
- Navigate to the indCAPS main page and examine the third blue box, labeled “Design of dCAPS Screening Primers for Isogenic Mutagenic Events”.
- Copy your wild-type and mutant sequences into the fields labeled “WT Sequence” and “Mutant Sequence”.
- Copy your CRISPR/Cas9 target sequence into the “CRISPR Target Sequence” field. This field has the same requirements as the corresponding field in the Screening Primers tool.
- Adjust parameters as necessary. The parameters are similar to those described by the CRISPR/Cas9 screening tool with the exception that no Acceptable Loss parameter is present.
- Press “Submit”.
- The output and selection criteria are the same as those described for the CRISPR/Cas9 screening tool.
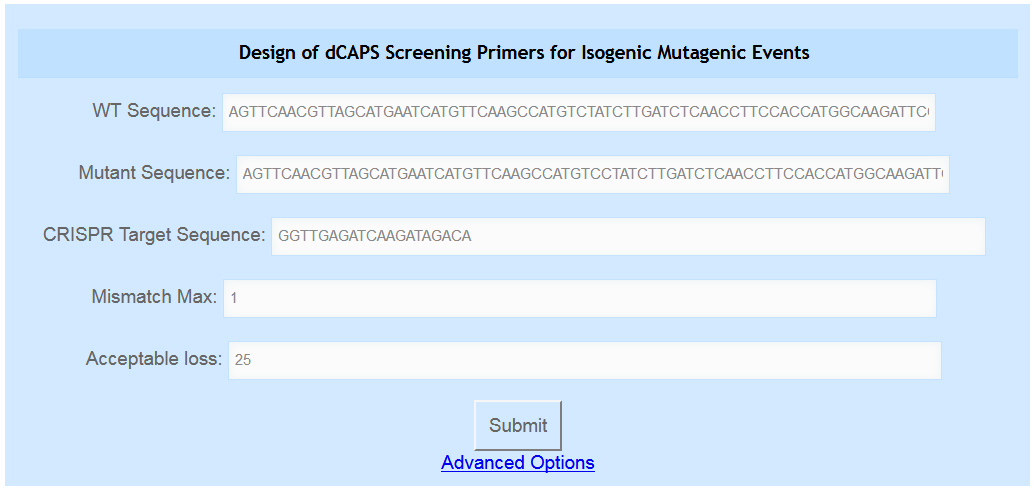
Figure 4. The user can enter their wild-type and mutant sequences in the first two text boxes and the CRISPR/Cas9 target sequence used in the third box. They can also indicate their desired maximum number of mismatches and acceptable loss parameter in the fourth and fifth boxes. Advanced options are accessed by clicking the blue “Advanced Options” link.
Data analysis
The algorithm used by indCAPS is described in “indCAPS: A tool for designing screening primers for CRISPR/Cas9 mutagenesis events” (Hodgens et al., 2017), available at https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0188406.
Acknowledgments
This work was supported by grants from the National Science Foundation (IOS-1238051 and IOS-1455607) as well as the USDA (2018-67013-27423).
Competing interests
The authors have no competing interests to declare.
References
- Hodgens, C., Nimchuk, Z. L. and Kieber, J. J. (2017). indCAPS: A tool for designing screening primers for CRISPR/Cas9 mutagenesis events. PLoS One 12(11): e0188406.
- Neff, M. M., Neff, J. D., Chory, J. and Pepper, A. E. (1998). dCAPS, a simple technique for the genetic analysis of single nucleotide polymorphisms: experimental applications in Arabidopsis thaliana genetics. Plant J 14(3): 387-392.
- Neff, M. M., Turk, E. and Kalishman, M. (2002). Web-based primer design for single nucleotide polymorphism analysis. Trends Genet 18(12): 613-615.
Article Information
Copyright
© 2019 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Hodgens, C., Nimchuk, Z. L. and Kieber, J. J. (2019). Using indCAPS to Detect CRISPR/Cas9 Induced Mutations. Bio-protocol 9(18): e3374. DOI: 10.21769/BioProtoc.3374.
Category
Molecular Biology > DNA > Genotyping
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








