- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
A Widely Applicable Urea-based Fluorescent/Colorimetric mRNA in situ Hybridization Protocol
Published: Vol 9, Iss 17, Sep 5, 2019 DOI: 10.21769/BioProtoc.3360 Views: 6875
Reviewed by: Gal HaimovichVarpu MarjomakiAnca Flavia Savulescu

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

APEX2 RNA Proximity Labeling in Mammalian Cell Lines With Low Biotin Permeability
Adrian Beat Tschan [...] Alexa B.R. McIntyre
Jul 5, 2025 2321 Views

Establishing and Maintaining 3D Organoid Cultures From Human Fallopian Tube Epithelium
David Holthaus [...] Thomas F. Meyer
Aug 20, 2025 2202 Views

Examining the Roles of m6A Sites in mRNA Using the Luciferase Gene Fused With Mutated RRACH Motifs
Nobuko Katoku-Kikyo and Nobuaki Kikyo
Nov 5, 2025 1938 Views
Abstract
In situ hybridization methods are routinely employed to detect nucleic acid sequences, allowing to localize gene expression or to study chromosomal organization in their native context. These methods rely on the pairwise binding of a labeled probe to the target endogenous nucleic acid sequence–the hybridization step, followed by detection of annealed sequences by means of fluorescent or colorimetric reactions. Successful hybridization requires permeabilization of tissues, followed by denaturation of nucleic acids strands, which is usually carried out in a formamide-based buffer and at high temperatures. Such reaction conditions, besides posing a health hazard (both concerning manipulation and waste disposal), can be excessively harsh for the delicate tissues of some species or developmental stages. We detail here an alternative method for in situ hybridization, where the toxic formamide is replaced with a urea solution. This substitution improved both tissues preservation and signal-to-noise detection, in several animal species. The protocol described here, originally developed for the hydrozoan jellyfish Clytia hemisphaerica, provides guidelines for adapting formamide-based traditional protocols to the urea variant. Urea-based protocols have already been successfully applied to diverse invertebrate and vertebrate species, showing the ease of such a modification, and providing the scientific community with a promising, safer and versatile tool.
Keywords: mRNA in situ hybridizationBackground
The pairwise complementary binding of single strand nucleic acid sequences inspired the development of a powerful technique, in situ hybridization, which allows researchers to visualize the location of DNA or RNA strands within their cellular and tissue contexts (Pardue and Gall, 1969). Numerous variants have since been developed, differing for the labeling of the exogenous complementary probes (e.g., radioactive or hapten-based), in the type of samples (e.g., for whole embryos or tissue sections) and in the detection method (e.g., fluorescent or colorimetric signals).
Precise annealing of probes depends on the accessibility of the target sequences, achieved through appropriate permeabilization of tissues and cell membranes (which allows penetration of reagents), and denaturation of nucleic acid strands (which exposes the complementary target sequences). Achieving adequate sensitivity of detection, specificity of annealing, and preservation of sample morphology often requires a time-consuming optimization of reaction conditions. Efficient denaturation and annealing of nucleic acids strands are usually obtained by means of elevated temperatures and the addition of a denaturing reagent in the hybridization buffer.
Reaction temperatures should ideally be adapted to the base composition of the sequences of interest: hybridization temperatures are about 25 °C below denaturation temperatures (Tm; Marmur and Doty, 1961). 50-70% formamide is usually employed as the denaturing reagent, this choice remaining largely unchallenged since the ‘80s–with few exceptions, such as the smFISH (single-molecule Fluorescence In Situ Hybridization) approach where 10-15% formamide is used (Haimovich and Gerst, 2018).
Elevated temperatures increase evaporation of the hybridization buffer, compounding the health risk already posed by formamide, an irritating, embryotoxic and teratogenic solvent (Gleich, 1974; Stula and Krauss, 1977; Merkle and Zeller, 1980; Kennedy and Short, 1986; Fail et al., 1998; George et al., 2000 and 2002; see also Sinigaglia et al., 2018 for further information). Hybridizing samples and waste need to be appropriately handled (CICAD 31), thus limiting the application of standard formamide-based protocols in sensitive contexts, such as pregnancy or in classrooms.
In the early years, a diversity of solvents was found to effectively destabilize nucleic acid strands, with urea being particularly efficient (Herskovits, 1963). Indeed, urea and formamide have similar properties, and are employed equivalently in a number of applications, such as fluorescent in situ hybridization on bacteria (Fontenete et al., 2016), protein denaturation, and clearing agents for imaging (reviewed in Azaripour et al., 2016). Urea can effectively replace formamide in Northern and Southern blot experiments, in particular at concentrations in the 2 M-4 M range (Simard et al., 2001). The mechanism of action of urea is still relatively poorly understood. Urea lowers the Tm of DNA of about 2 °C per mole of urea (Hutton, 1977), and is thus slightly less efficient than formamide, which achieves a 2.4-2.9 °C reduction per mole. Urea molecules bind weakly to RNA, disrupting its base-pair interactions and ultimately destabilizing the structure of RNA molecules (Herskovits and Bowen, 1974; Priyakumar et al., 2009; Lambert and Draper, 2012).
Urea, in powder or crystal form, is water soluble, and 8 M stock solutions can be readily prepared at 20 °C. Solutions tend to be viscous (Hutton, 1977), which might explain why high concentrations of urea are less effective (Simard et al., 2001). Such increased viscosity demands care during the hybridization step of the in situ protocol, to ensure that samples are efficiently immersed, and evaporation minimized.
The urea-based protocol presented here has been shown to be more efficient than the standard formamide-based ones in diverse metazoan species and at different developmental stages (see Sinigaglia et al., 2018). Detection sensitivity was improved, allowing visualization of unsuspectedly complex patterns of gene expression (Sinigaglia et al., 2018). This increased sensitivity of detection might be explained by an additional permeabilizing action of urea on tissues (Lim et al., 2009; Huang et al., 2011), likely due to increased osmotic pressure within cells (reviewed in Tainaka et al., 2016). The hyperhydration of tissues might also partially account for the improved morphologies of delicate samples, shown in the original research paper (Sinigaglia et al., 2018).
The protocol detailed here was originally developed for the hydrozoan medusa Clytia hemisphaerica (Quiroga Artigas et al., 2018; Sinigaglia et al., 2018, Leclère et al., 2019). Following those guidelines, urea-alternative protocols have already been developed for a diversity of metazoan species and developmental stages, including priapulid and brachiozoan embryos (Thiel et al., 2017; Sinigaglia et al., 2018), the acoel Hofstenia miamia (L. Ricci, personal communication), the scyphozoan jellyfish Aurelia aurita (M. Manuel and T. Condamine, personal communication), and in the paddlefish (M. Minarik, personal communication). Substituting formamide with urea has also been applied to DNA-FISH on mouse oocytes (Manil-Segalen et al., 2018), further showing the versatility of this approach.
Such versatility stems from the simplicity of the key implementation, the substitution of formamide with an equal volume of urea solution (at a final concentration of 4 M). When adapting previous protocol to novel species or tissues, it is recommended to start by simply switching formamide to urea, leaving the rest of the procedure unchanged. Guidelines for eventual troubleshooting are also provided, offering general recommendations for successful in situ hybridization.
Materials and Reagents
- Materials and reagents used in multiple steps
- Gloves
- RNase free tubes (e.g., 1.5 ml Eppendorf tubes, or equivalent)
- In situ baskets (Sample container, as in Figure 1A)
They could be either the Netwell baskets (Corning), or homemade baskets constructed from nylon mesh and microcentrifuge tubes, as explained in Sive et al. (2007). Baskets size depends on the type of sample; they usually fit within the wells of a 6-, 12- or 24-well plate. Performing in situ hybridization in baskets (instead of tubes) is recommended:- To reduce loss and damages to samples, due to pipetting.
- To accelerate exchange of solutions (baskets can be simply transferred with the help of forceps to a multiwell plate with the new solution (Figure 1B)–in this case tap gently the basket against the walls of the well to ensure removal of older solution, then gently immerse it in the new well).
- Baskets can be recycled multiple times, after being thoroughly washed (e.g., rinse with MilliQ H2O, then immerse in a 1 M NaOH solution to neutralize RNases, and rinse several times with RNase-free MilliQ H2O–simply placing them in a beaker on a shaker will assure thorough swirling).

Figure 1. Sample carrier. A. Example of three baskets with mesh-bottom. B. A 24-well plate used for in situ hybridization, with four baskets. C. The same plate, closed and wrapped in plastic film to avoid evaporation during hybridization step.
- Multiwell plate (For fitting in situ baskets, as in Figure 1B); (e.g., 24-well cell culture plate, sterile, with lid, from CellStar (Greiner Bio-one, catalog number: 662-160))
- Ice and ice container
- MilliQ H2O (Storage: RT)
- Tween-20 (Sigma-Aldrich, catalog number: P1379), storage: RT (Mild detergent. Given its high viscosity, it is recommended to prepare a 10% working solution)
- Na2HPO4·7H2O (Sigma-Aldrich, catalog number: 431478), storage: RT (For preparing 10X PBS solution)
- NaCl (Sigma-Aldrich, catalog number: S9888), storage: RT (For preparing 10X PBS solution)
- KCl (Sigma-Aldrich, catalog number: P9333), storage: RT (For preparing 10X PBS solution)
- KH2PO4 (Sigma-Aldrich, catalog number: P9791), storage: RT (For preparing 10X PBS solution)
- 10x PBS (Phosphate-Buffered Saline) pH 7.4 (see Recipes, or from tablets from Calbiochem, catalog number: 524650), storage: RT (Helps maintaining a constant pH)
- Fixation
- 37% formaldehyde (Sigma-Aldrich, catalog number: 252549), storage: RT (Crosslinking)
- 8% glutaraldehyde (Electron Microscopy Sciences, catalog number: 16020), storage: 4 °C (Crosslinking)
- EGTA (Ethylene glycol-bis(2-aminoethylether)-N,N,N',N'-tetraacetic acid) (Sigma-Aldrich, catalog number: E3889), storage: RT [Calcium/magnesium chelator, which reduces enzyme activity (such as RNase); reagent of HEM buffer]
- HEPES (Sigma-Aldrich, catalog number: H4034), storage: RT (Reagent of HEM buffer)
- MgSO4 (Sigma-Aldrich, catalog number: M7506), storage: RT (Reagent of HEM buffer)
- Tween-20 (Sigma-Aldrich, catalog number: P1379), storage: RT
- Methanol (e.g., Sigma-Aldrich, catalog number: 322415), storage: RT
- 1x PBS (see Recipes)
- 1x PBST (see Recipes)
- CISH fixation buffer (see Recipes)
- FISH fixation buffer (see Recipes)
- Dehydration buffer (see Recipes)
- Rehydration buffer (see Recipes)
- Re-fixation buffer (see Recipes)
- Acetylation
- TEA (Triethylamine) (Sigma-Aldrich, catalog number: T0886), storage: RT (For reducing the non-specific binding of negatively-charged probe to tissues)
- Acetic anhydride (Sigma-Aldrich, catalog number: A6404), storage: 4 °C (For reducing the non-specific binding of negatively-charged probe to tissues)
- 1x PBST (see Recipes)
- 10x TEA stock solution (see Recipes)
- 0.1 M TEA (see Recipes)
- 0.25% Acetic anhydride in 0.1 M TEA (see Recipes)
- Permeabilization
- Proteinase K (Sigma-Aldrich, catalog number: P4850), storage: 4 °C or -20 °C (Digests proteins, including nucleases. Optimum pH is 7.5-9.0)
- Glycine (Sigma-Aldrich, catalog number: G8898), storage: RT (For neutralizing the proteinase K)
- 1x PBST (see Recipes)
- Hybridization
- Plastic film (For wrapping sample container during hybridization step)
- Urea (Sigma-Aldrich, catalog number: U5378), storage: RT (Denaturing agent)
- Dextran sulfate sodium salt, Mr ~200''00 (Sigma-Aldrich, catalog number: 67578-25G), storage: RT [Increases molecular crowding and can therefore accelerate the kinetics of hybridization (locally increases probe concentration)]
- tRNA, from baker’s yeast (Sigma-Aldrich, catalog number: 10109509001), storage: -20 °C (Blocks non-specific probe binding)
- Salmon sperm (Thermo Fisher, catalog number: 15632011), storage: -20 °C (Blocks non-specific probe binding)
- Heparin sodium (Thermo Fisher, catalog number: 10239840), storage: -20 °C (Helps to reduce background, by binding to proteins that bind to DNA)
- 20% SDS (Sodium Dodecyl Sulfate) (Fisher, catalog number: BP131), storage: RT (Detergent. Permeabilizes membranes, facilitating probe distribution)
- NaCl (Sigma-Aldrich, catalog number: S9888), storage: RT (Salt. For preparing 20x SSC stock buffer, see Recipes)
- Trisodium citrate (Na3C6H5O7) (For preparing 20x SSC stock buffer, see Recipes)
- Tween-20 (Sigma-Aldrich, catalog number: P1379), storage: RT
- 10x DIG labeling mix RNA (Roche, catalog number: 11277073910), storage: -20 °C (For probe synthesis)
- RNasin Ribonuclease Inhibitor (Promega, catalog number: N2111 or N2511), storage: -20 °C (For probe synthesis)
- DTT (Promega, catalog number: P1171), storage: -20 °C (For probe synthesis)
- Transcription Optimized 5x Buffer (Promega, catalog number: P1181), storage: -20 °C (For probe synthesis)
- T7/SP6/T3 RNA polymerase (Promega, catalogue number, respectively: P2075/ P1085/ P2083), storage: -20 °C (For probe synthesis)
- RQ1 RNase-free DNase (Promega, catalog number: M6101), storage: -20 °C (For probe synthesis)
- Illustra ProbeQuant G-50 Micro Columns (GE Healthcare, catalog number: 28-9034-08) (For probe synthesis)
- 1x PBST (see Recipes)
- 8 M Urea stock solution (see Recipes)
- 20x SSC (Sodium Saline Citrate or Standard Saline Citrate) (see Recipes, or from Sigma-Aldrich, catalog number: S6639), storage: RT [Controls stringency (annealing of probe to target)]
- Wash I (see Recipes)
- Wash II (see Recipes)
- Wash III (see Recipes)
- Urea-based 100% Hybridization Buffer (see Recipes)
- Urea-based 50% Hybridization Buffer (see Recipes)
- Antibody incubation
- Anti-Digoxigenin-AP, Fab fragments (Roche Applied Sciences, Sigma-Aldrich, catalog number: 11093274910), storage: -20 °C [Recognizes the DIG hapten, conjugated to Alkaline Phosphatase (AP)]
- Anti-Digoxigenin-POD, Fab fragments (Roche Applied Sciences, Sigma-Aldrich, catalog number: 11207733910), storage: -20 °C [Recognizes the DIG hapten, conjugated to horseradish Peroxidase (POD)]
- Blocking reagent (Sigma-Aldrich, catalog number: 11096176001), storage: 20-25 °C (To decrease non-specific binging of antibodies)
- Maleic acid (Sigma Aldrich, catalog number: M0375), storage: RT (For preparing MAB)
- NaCl (Sigma-Aldrich, catalog number: S9888), storage: RT (Salt. For preparing MAB)
- NaOH (Sigma-Aldrich, catalog number: 221465), storage: RT (For preparing MAB)
- 10x blocking reagent stock solution (see Recipes), storage: -20 °C
- 10x MAB (Maleic Acid Buffer) stock solution (see Recipes) (For diluting blocking buffer, used after SSC washes)
- 1x MABT buffer (see Recipes)
- 10x blocking reagent stock solution (see Recipes), storage: -20 °C
- 1x Blocking buffer (see Recipes)
- Detection
- Aluminum foil (For protecting samples during staining step)
- BCIP (5-Bromo-4-Chloro-3-Indolyl-Phosphate)/NBT (Nitro Blue Tetrazolium) Color Development Substrate (Promega, catalog number: S3771), storage: 4 °C or -20 °C [Substrate of alkaline phosphatase which generates a dark (blue/violet) precipitate]
- NaCl (Sigma-Aldrich, catalog number: S9888), storage: RT (Salt. Component of NTMT buffer)
- MgCl2 (Sigma-Aldrich, catalog number: M8266), storage: RT (Component of NTMT buffer, MgCl2 is a necessary cofactor for alkaline phosphatase)
- Tris-HCl (Tris hydrochloride) (Sigma, catalog number: T3253), storage: RT (pH buffering component of NTMT)
- Tween-20 (Sigma-Aldrich, catalog number: P1379), storage: RT
- Levamisole (Agilent Dako, catalog number: X302130-2), storage: 2-8 °C (Alkaline Phosphatase inhibitor)
- TSA Plus Fluorescence Amplification kit Cyanine 3/5 (Perkin Elmer, Waltham, MA; catalog number: NEL752001KT), storage: 4 °C [Tyramide Signal Amplification (TSA) kit uses Horseradish Peroxidase (HRP or Horseradish POD) to catalyze the deposition of a fluorophore-labeled radicals]
- H2O2 (Sigma-Aldrich, catalog number: 516813), storage: 4 °C (Substrate for horseradish peroxidase)
- Filters for syringe (Millex-GS, catalog number: SLGS033SS) (For filtering NTMT buffer, in order to avoid precipitates)
- Disposable syringes (e.g., 50 ml from Thermo Scientific, catalog number: S7510-50) (For filtering NTMT buffer, in order to avoid precipitates)
- 5 M NaCl stock solution (For preparing NTMT buffer, see Recipes), storage: RT
- 1 M Tris-HCl, pH 9.5 stock solution (For preparing NTMT buffer, see Recipes), storage: RT
- 1 M MgCl2 stock solution (For preparing NTMT buffer, see Recipes), storage: RT
- NTMT buffer (see Recipes)
- NTMT minus buffer (see Recipes)
- NBT/BCIP in NTMT buffer (see Recipes)
- Storage & Mounting
- Glycerol (Sigma-Aldrich, catalog number: G5516), storage: RT (Mounting medium)
- Citifluor-AF1 (Citifluor, catalog number: AF1-100), storage: RT [Mounting medium for fluorescent samples (on-hardening antifading/anti-bleaching)]
- Slide (e.g., 76 x 26 mm, with frosted end (practical for labeling), from Knittel Glass (Mounting)
- Coverslide (e.g., 20 x 20 mm, thickness 1, from Knittel Glass) (Mounting)
- Nailpolish (For sealing mounted specimens; any commercial nailpolish can be used, however prefer a transparent one, and test first on lesser important samples, in case it contains too much solvent–which might damage samples)
Equipment
Note: No specific brand/model is provided; this equipment is commonly available in laboratories.
- Pipettes
- Heating block (For probe denaturation, e.g., featuring a temperature range 10-100 °C)
- Metallic forceps (For transferring baskets, e.g., from Fine Science Tools)
- Waterbath/Hybridization oven (Hybridization and stringent washes)
- Glass bottles with high-temperature resistant screw caps (e.g., 250 ml bottle from Fisherbrand (catalogue number: FB-800-250); 500 ml bottle from VWR [borosilicate 3.3; catalogue number: 215-1594); 1000 ml bottle from Scott Duran (Original GL 45 series)]
- pH meter (For adjusting pH of solutions)
Procedure
General notes on samples handling:
- Sample fixation is best performed in tightly sealed RNase free containers, such as Eppendorf or Falcon tubes.
- Once samples are re-hydrated, they can be transferred to RNase free in situ hybridization baskets (shown in Figure 1A).
- The density of samples might affect the in situ hybridization results: overfilled vials/baskets often yield poorly stained/damaged samples, and too little amount of sample (e.g., one embryo) might become over-stained (possibly due to unbalanced ratio probe/target).
- Sample fixation/dehydration (Table 1)
Notes:
- Fixatives should be handled and disposed of according to laboratory safety guidelines.
- Fixation (choice of fixative, duration of fixation) must be adapted to sample types.
- Two alternative types of fixation are provided, for colorimetric in situ hybridization (CISH), and for fluorescent in situ (FISH).
Table 1. Fixationa

- Rehydration (Table 2)
Table 2. Rehydration
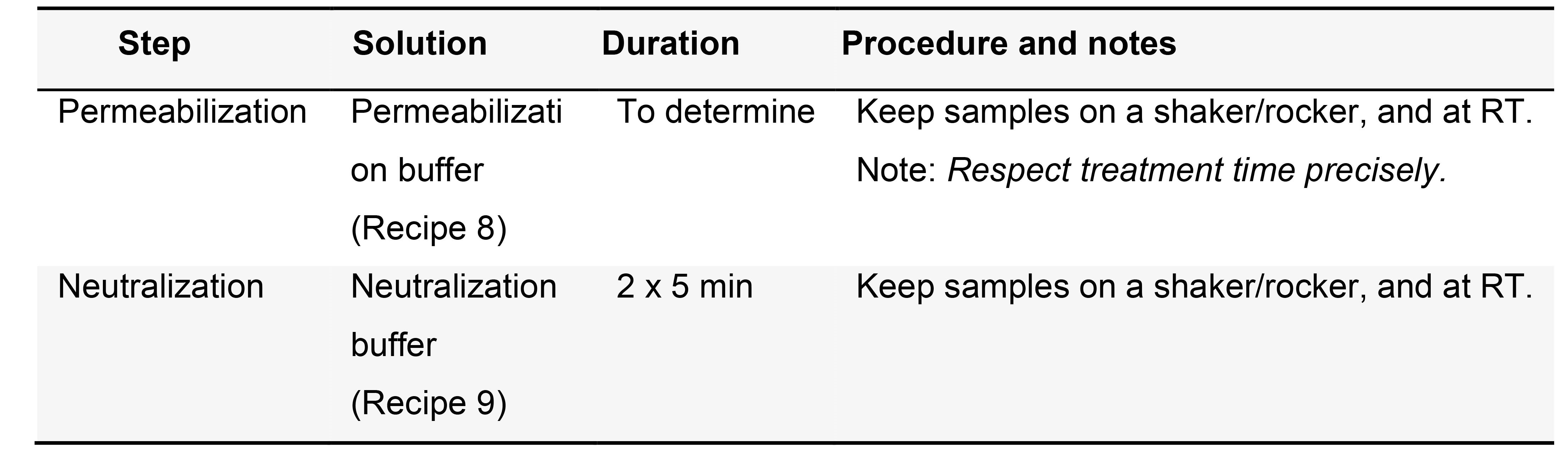
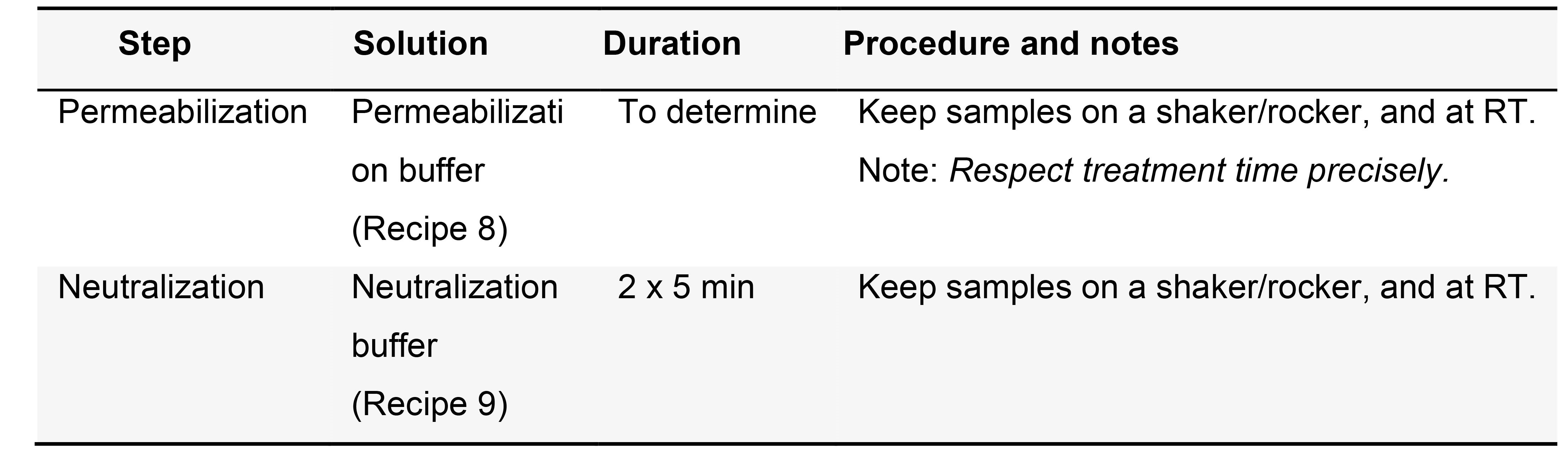
- (Optional) Permeabilization; proteinase K treatment permeabilize tissues (Table 3)
Notes:- Treatment time should be titrated for each new batch of enzyme, as strength might vary. A useful rule of thumb is treating a test sample until tissue damage appears, and halving the resulting time (see Figure 3 in Notes section for further details).
- This step might prove unnecessary for very delicate tissues.
- (Optional) Acetylation (Table 4)
Note: This step might help reducing non-specific signal, due to probes binding to positively charged surfaces.
Table 4. Acetylation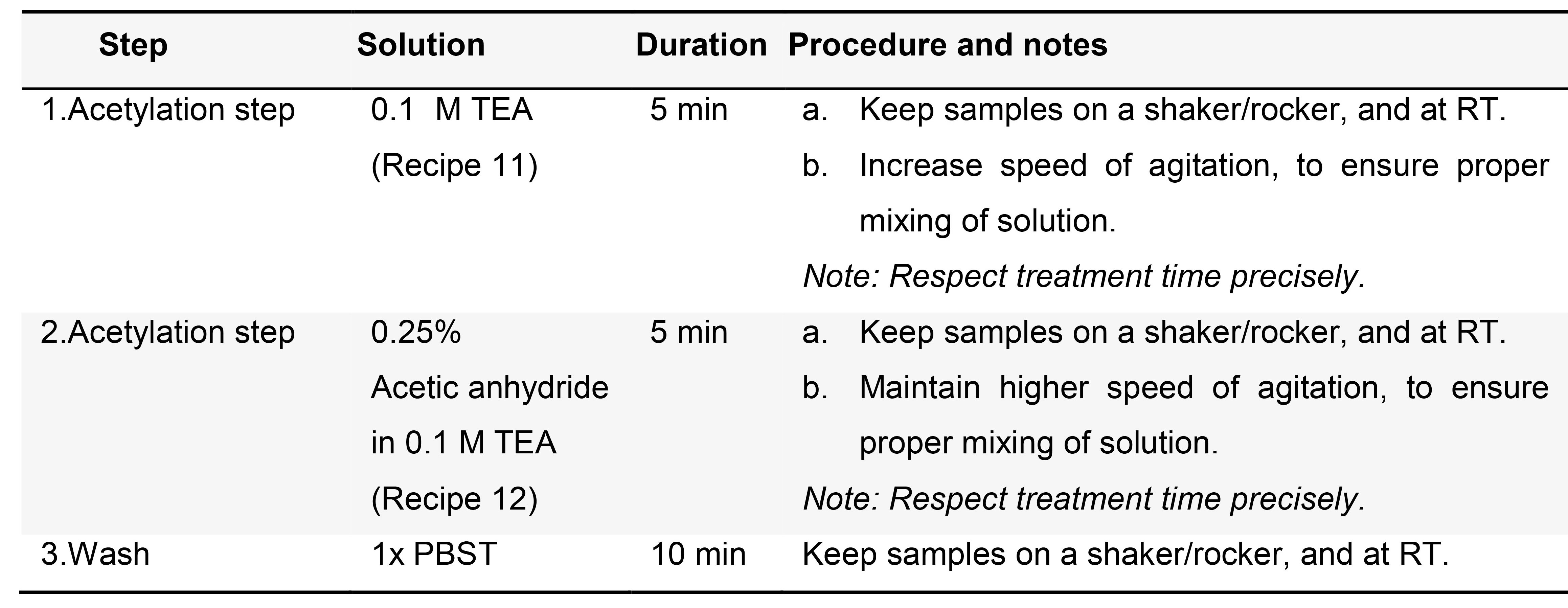
- (Optional) Re-fixation (Table 5)
Notes:
- Depending on sample type (e.g., fragile invertebrate embryos), an additional re-fixation step prior to hybridization might be required.
- Generally, it is advisable to re-fix after proteinase K treatment.
- Glutaraldehyde might be omitted: it is recommended to test several options while setting up the protocol for the first time. Glutaraldehyde penetrates tissues slowly, and its use is preferable for samples thinner than 1 mm (Eltoum et al., 2001).
Table 5. Re-fixation
- Hybridization (Table 6)
Notes:
- Do not let the temperature drop during the pre-hybridization/hybridization phase. Transfer of baskets or addition of probe might take long time. When working on multiple samples, set up manageable batches.
- The urea-based hybridization buffer tends to be viscous, and evaporation can be an important issue, leading to the formation of crystals that will damage the samples. Seal vials/multi-well plate carefully (Figure 1C).
- Be aware that some sample types might become temporarily transparent while in heated hybridization buffer. In this phase, they will be also more fragile.
Table 6. Hybridization
- Blocking (Table 7)
Table 7. Blocking
- Signal detection
AP antibody, colorimetric reaction (Table 8)
Table 8. Colorimetric signal detection

Fluorescent detection (POD antibody, signal amplification) (Table 9)
Table 9. Fluorescent signal detection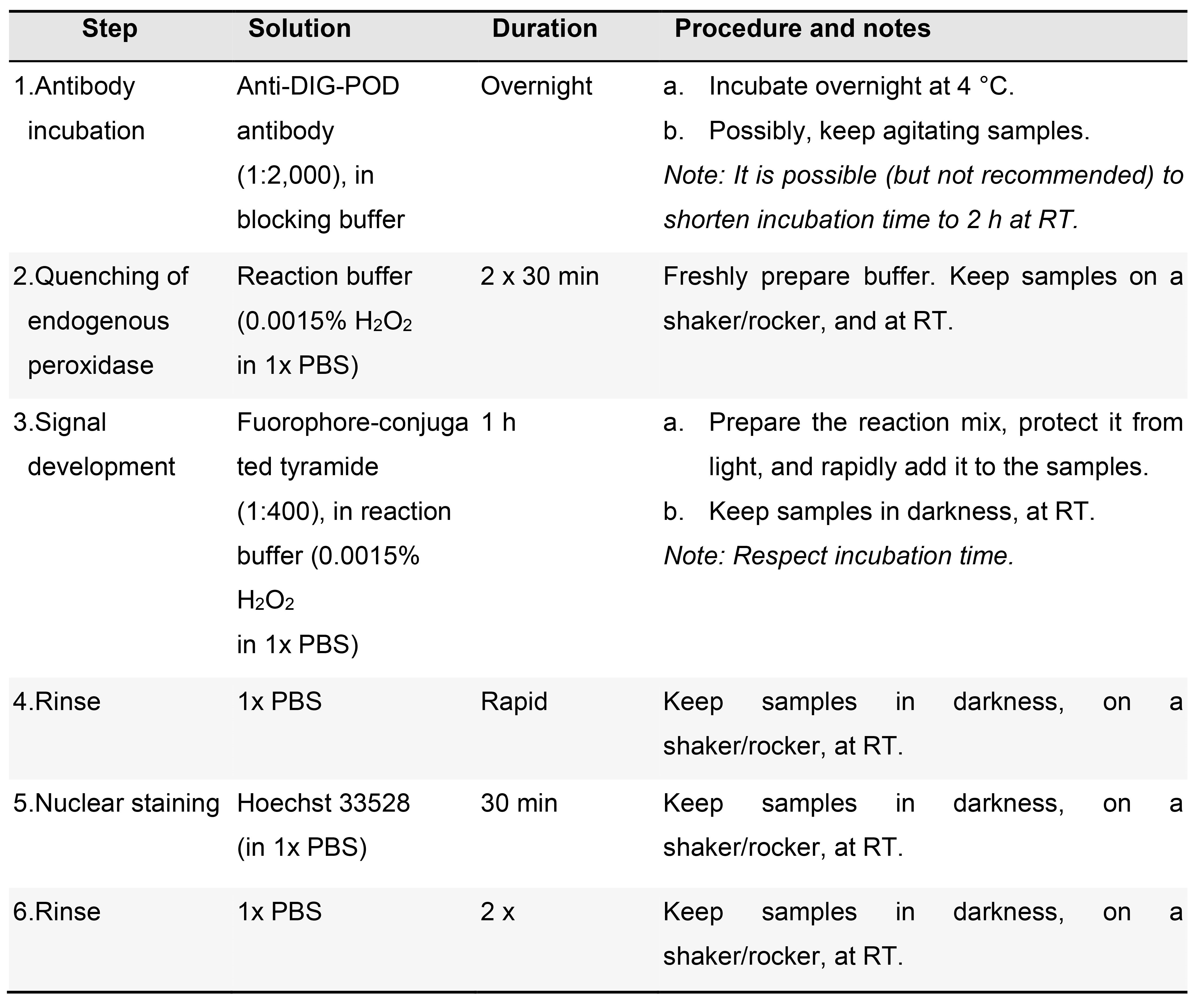
- CISH mounting (Table 10)
Table 10. Colorimetric samples- mounting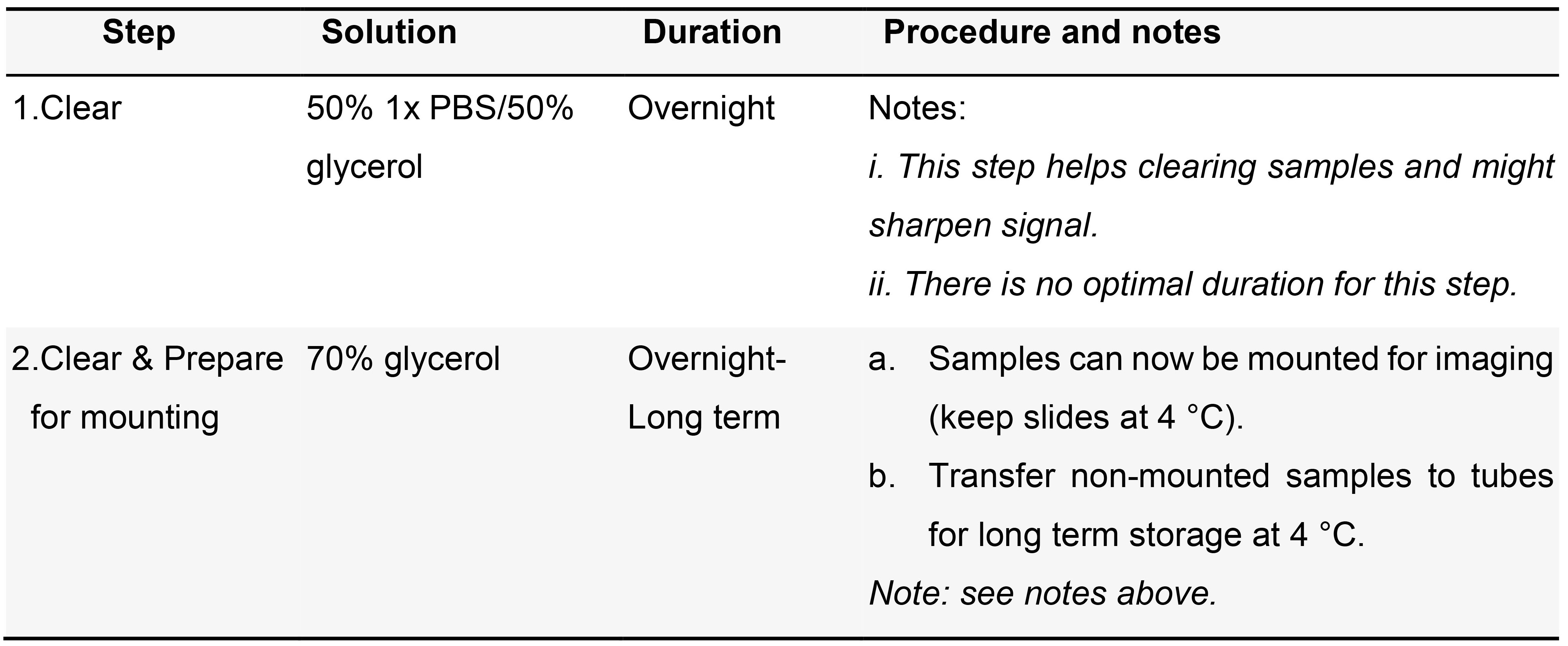
- FISH mounting (Table 11)
Table 11. Fluorescent samples- mounting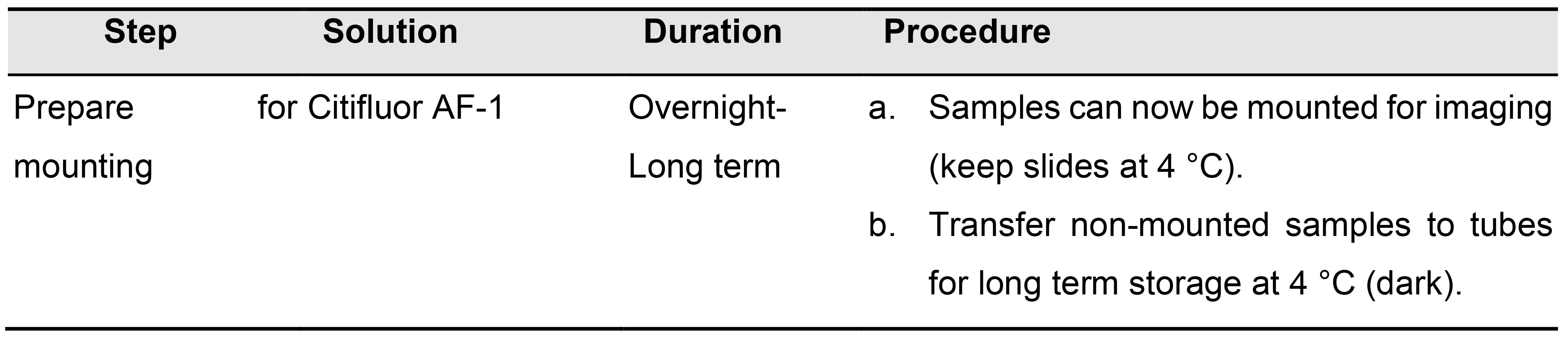
Data analysis
The urea-variant in situ hybridization protocol generates reliable results. Sample morphology is well preserved, and the good ratio signal-to-background allows for detecting of gene expression with cellular resolution. Figure 2 depicts the results of colorimetric (CISH) and fluorescent (FISH) protocols performed on a Clytia medusa.
Please refer to the original paper (Sinigaglia et al., 2018) for a further comparison of the formamide-based and urea protocol, for examples from diverse species, and for a more detailed analysis of results.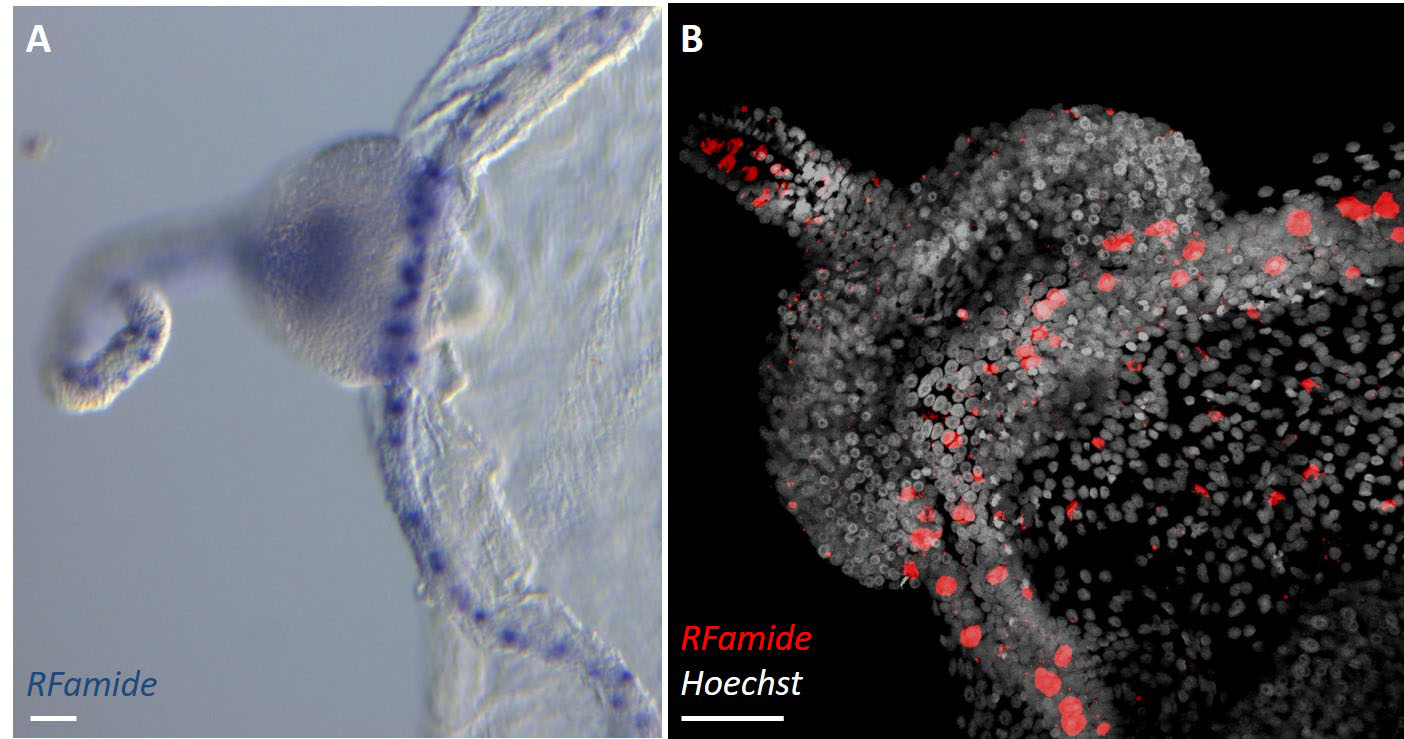
Figure 2. Detection of gene expression in medusae. RFamide (accession number for Clytia gene: KX496951) is expressed in nervous system cells. The urea- in situ hybridization protocol detects cells RFamide+, scattered thorough the medusa body; here a detail is shown, including (left to right): tentacle, tentacle bulb, circular canal running at the periphery of the umbrella, and umbrella. Tissues are well preserved, in particular the fragile umbrella. A. CISH result, the focus is on the scattered cells in the tentacle and in the circular canal. Image taken with a Zeiss Axio Imager A2. B. FISH results, maximum projection (done with Fiji) of a z-stack image taken with a Leica SP8 confocal microscope. Both images were cropped; scale bars = 50 μm.
Notes
- Controls
- It is highly recommended to perform both a negative control (e.g., using sense probes, or a probe of similar length for a gene not expressed in the sample being studied), and a positive control (using a reliable, known antisense probe), in particular when testing any new probe or optional step.
- When testing for the first time this urea-based protocol, it is recommended to perform a parallel in situ hybridization with an eventual previous formamide-based protocol (if one is already in use in the laboratory).
- mRNA probes
- Riboprobes (mRNA probes) can be synthesized from either a linearized template plasmid, or from a PCR product. The DIG-labeled RNA probes shown in the original research paper (Sinigaglia et al., 2018), including the RFamide probe employed in the present protocol, were synthesized using one of the RNA polymerase kits from Promega (T7, SP6 or T3 polymerases, according to the orientation of insert). For probe synthesis, add in order: 2 μl of 10x DIG labeling mix RNA (Roche); 0.5 μl of RNasin Ribonuclease Inhibitor (Promega); 2 μl of 100 mM DTT (Promega); 4 μl of Transcription Optimized 5x Buffer (Promega); 1-2 μg of DNA template (calculate the appropriate volume); RNase-free H2O to a 20 μl of total volume; 0.5 μl of T7/SP6/T3 RNA polymerase (Promega). Run reaction for 2-5 h at 37 °C, then arrest by digesting the DNA template with 1.5 μl of RQ1 RNase-free DNase (Promega), at 37 °C for 30 min. Elute by adding 30 μl of RNase free H2O, purify with the illustra ProbeQuant G-50 Micro Columns (GE Healthcare). Check probe integrity by running 1 μl of purified probe on 1% agarose gel, and quantify with nanodrop. Probes can be stored at -20 °C or -80 °C, addition of 50% (v/v) RNase-free formamide helps preserving RNA integrity.
- It is recommended to synthesize always both the antisense (complementary to mRNA) probe and the sense probe, to be used as a control for the specificity of binding.
- Probes are usually designed on the coding sequence of a gene, but they can also target the 3' UTR region (this might be helpful for example when discriminating highly similar genes). Clone the desired region from cDNA, possibly deriving from mRNA extracted from the stage/body region of interest (different stages/tissues might express differently-spliced forms of the same gene).
- Recommended probe length ranges between 500 and 800, up to 1,500 bp. Long probes can be fragmented, e.g., by alkaline hydrolysis.
Alkaline hydrolysis (Cox et al., 1984): Hydrolyze probe by adding 1:1 volume of hydrolysis buffer (40 mM NaHCO3/60 mM Na2CO3, pH 10.2) freshly prepared in RNase-free water. Incubate at 60 °C, for a length of time (expressed in minutes) calculated as: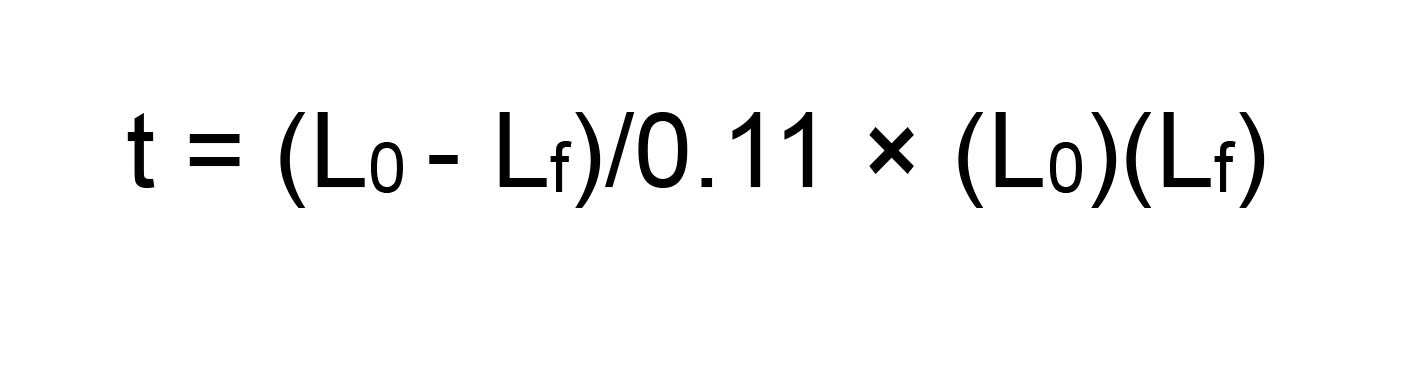
Where, L0 is original transcript length (expressed in kb) and Lf is the desired fragment length (in kb).
Neutralize by adding sodium acetate (to 0.1 M final, at pH 6.0), and glacial acetic acid (to 0.5%, v/v), and precipitate with ethanol.
- Adapting the protocol/Variants
- This protocol is based on Clytia in situ hybridization (Sinigaglia et al., 2018); it is however meant to represent a general guideline. When adapting it to new study organisms/tissues/cells, it is recommended to start by simply substituting the formamide in the original hybridization buffer, with a urea solution, to a final concentration of 4 M.
- Variants to the protocol are possible, in particular the fluorescent in situ hybridization has already been successfully coupled to immunostaining or to EdU reactions (data not shown).
- This protocol might be used also on tissue sections (frozen or paraffin-embedded), in which case further adjustments might be necessary (e.g., reducing detergents or urea concentration in the hybridization buffer).
- Urea-based hybridization buffer might also be tested with smFISH.
- Proteinase K treatment is optional. Proteinase K digests proteins, thus permeabilizing tissues, and aiding the penetration of probes and reagents. Too strong treatment might damage tissue morphology, while insufficient digestion might result in poor detection of signal. Both incubation time and enzyme concentration should be assessed for each new batch of enzyme.
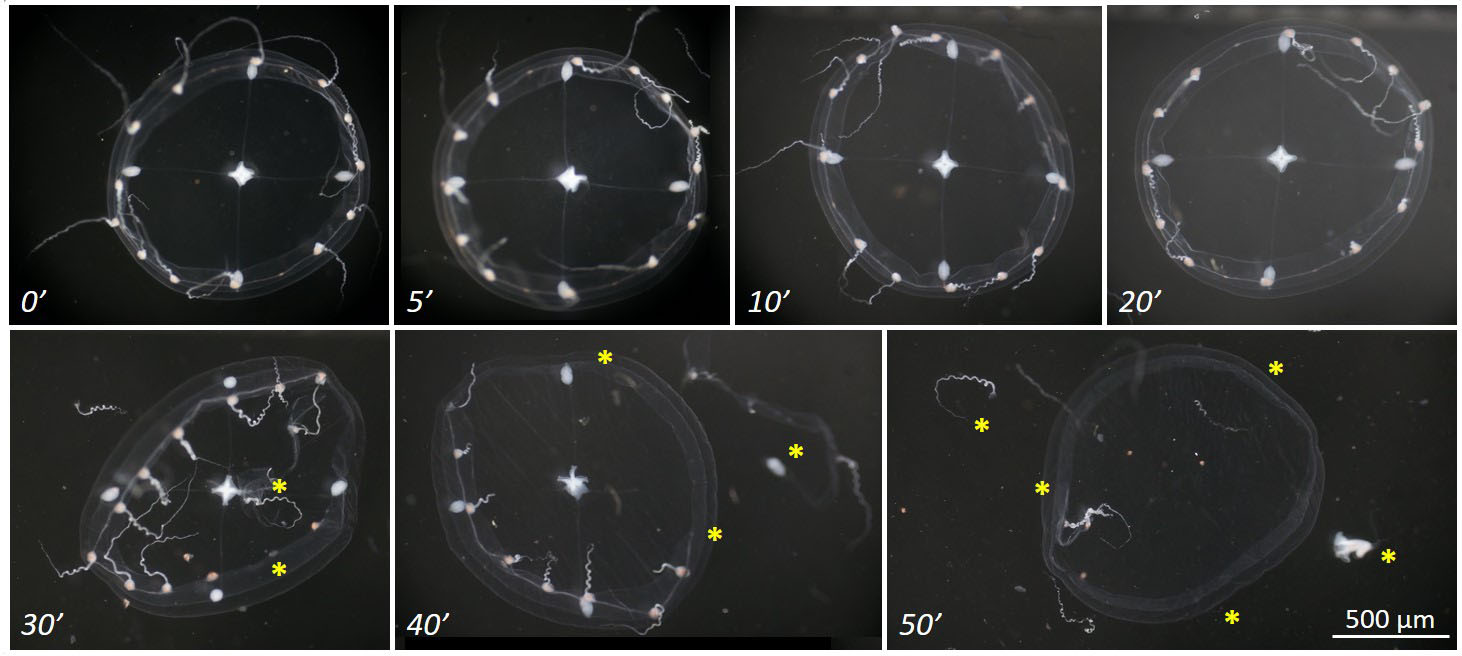
Figure 3. Titration of proteinase K treatment. Images show the progressive digestion of a fixed Clytia medusa, treated with 10 μg/ml of proteinase K in 1x PBST. Visible damages (yellow asterisk) start to appear after 30 min of treatment, and by 40 min the medusa is severely damaged (notice the detached velum and gonad). According to the guideline provided in the main protocol (see Table 3), the proteinase K treatment should last for 15-20 min during in situ hybridization.
- Troubleshooting
- Damaged samples
- Make sure that samples are always covered by solution, and never let them dry.
- Verify that hybridization buffer did not evaporate.
- Add a second fixation step prior to hybridization, after rehydration.
- Remove permeabilization step, or eventually reduce duration of treatment.
- Handle samples and baskets carefully. Samples might be more fragile while in methanol and in hybridization solution.
- If damages are due to crystals forming on samples, try rapidly washing samples with MilliQ H2O.
- Absent/poor signal
- Verify probe quality (check for degradation) and that correct antisense probe is being used. Long probes usually perform worse, the optimal length is however species- and gene-specific (aim for 500-800 bp). Probes can be fragmented (e.g., by alkaline hydrolysis, see point B4 of Notes section); this will improve both penetration and recognition of target.
- Add permeabilization step/re-test/renew proteinase K batch.
- Increase/reduce probe concentration in hybridization mix.
- In case of CISH: extend the signal detection step (renew frequently the NBT/BCIP mix, while keeping samples at 4 °C). In case of FISH: extend the signal detection reaction, or repeat it.
- In case of CISH: use freshly prepared staining solution. Freshly prepare also NTMT (do not use if older than a day, keep at 4 °C while not in use).
- In case of FISH: prepare buffer just before use.
- Increase/reduce urea concentration in the hybridization mix (8 M to 2 M): in general, higher concentrations will have a stronger permeabilizing effect, and are recommended for difficult/hard tissues.
- Verify the pH of solutions, in particular of TEA and of SSC stock solutions.
- Samples might be over-fixed, verify. It might be advisable to remove glutaraldehyde, in case it was added to the fixative solution.
- Non-specific signal
- In case of CISH: add levamisole to NTMT and staining mix.
- In case of CISH: extend incubation time with NTMT, prior to NBT/BCIP color reaction.
- In case of CISH: protect NBT/BCIP mix from light. Verify that the stock solution did not turn purple or black and that it appears homogeneous.
- In case of CISH: try performing FISH instead (allows for more precise observation, avoids eventual issues with non-specific deposition of precipitate).
- Pre-hybridize overnight.
- Add acetylation step.
- Make sure that acetic anhydride solution (Steps 1 and 2 in Procedure D) gets efficiently mixed, and that samples get well in contact with it.
- Increase hybridization temperature.
- Reduce probe concentration. Recycle probe (store the Hybridization mix at -20 °C).
- Increase duration/stringency of post-hybridization washes. Stringency is modulated by changing the concentration of SSC buffer (5-6x for low stringency, 2-0.1x for high stringency).
- Make sure that no dust/dirt is attached to specimens before starting the color reaction: particles might derive from insufficient washing of recycled baskets, from solutions (filter them, in particular NTMT solution), from clothes. If present, remove them manually.
In general, particles attached to specimens most commonly derive from old solutions: renew them, in particular the staining reagents, and adjust pH accurately. A step in NTMT minus buffer (filtered) prior to staining might also help - In case of CISH: extend Steps 9 in Procedure H (Colorimetric detection) and Procedure I (CISH mounting), keeping samples at 4 °C.
- Verify the pH of solutions, in particular of TEA and of SSC stock solutions.
- Signal too strong/rapid
- Reduce probe concentration.
- For CISH: dilute staining solution. Reduce staining time. Add levamisole to staining solution and/or carry out staining reaction at 4 °C, in order to slow down AP reaction.
- In case of CISH: try performing FISH instead (allows for more precise observation, avoids eventual issues with non-specific deposition of precipitate).
- Increase the stringency of washes.
- Add acetylation step.
- Damaged samples
Recipes
- 10x PBS (pH 7.4) stock solution
- Weigh and add: 25.6 g of Na2HPO4·7H2O, 80 g of NaCl, 2 g of KCl, 2 g of KH2PO4
- Bring to 1 L with MilliQ H2O
- Autoclave
- Store at room temperature, discard if precipitate/particles appear (the bottle might have been contaminated)
- 1x PBST
- Add 0.1% Tween-20 to 1x PBS solution
- Store at room temperature, discard if precipitate/particles appear (the bottle might have been contaminated)
- HEM buffer
0.1 M HEPES, pH 6.9
50 mM EGTA, pH 7.2
10 mM MgSO4
Prepare freshly - CISH fixation buffer
3.7% formaldehyde plus
0.4% glutaraldehyde
1x PBS (pH 7.4)
Prepare freshly - FISH fixation buffer
3.7% formaldehyde in HEM buffer
Prepare freshly - Dehydration buffer
50% 1x PBST
50% Methanol
Prepare freshly - Rehydration buffer
50% Methanol
50% 1x PBST
Prepare freshly - Permeabilization buffer
10 μg/ml proteinase K in 1x PBST
Prepare freshly - Neutralization buffer
2 mg/ml glycine in 1x PBST
Prepare freshly - 10x TEA stock solution
- Prepare 1 M solution in H2O
- Adjust the pH to 7.8
- Do not autoclave
- Handle TEA with care (do not shake)
- Store at room temperature, discard if precipitate/particles appear (the bottle might have been contaminated)
- 0.1 M TEA
Dilute 10x TEA stock solution with 1x PBST, to 0.1 M Triethanolamine (TEA)
Prepare freshly - 0.25% Acetic anhydride in 0.1 M TEA
Add 0.25% acetic anhydride (v/v) to 0.1 M TEA solution, and mix thoroughly
Prepare freshly - Re-fixation buffer
3.7% formaldehyde plus 0.2% glutaraldehyde in 1x PBST
Prepare freshly - 8 M Urea stock solution
- Dissolve urea in MilliQ H2O, calculating the required amounts (MW = 60.06 g/mol)
- Be aware that the reaction is endothermic
- Prepare freshly and sterilize by filtration if necessary
- The solution can be used for both hybridization and post-hybridization steps, in this case keep at 4 °C
- 20x SSC (sodium saline citrate) stock buffer (1 L)
Note: 20x concentration of buffer prevents fungal and bacterial growth.- Dissolve 175.3 g NaCl and 88.2 g of trisodium citrate (Na3C6H5O7) in 900 ml MilliQ H2O
- Adjust the pH to 7.0, top to 1 L with MilliQ H2O
- Autoclave to sterilize
- Store at room temperature, discard if precipitate/particles appear (the bottle might have been contaminated)
- 20% SDS (Sodium Dodecyl Sulfate)
Precipitation can occur at low temperatures. In this case, just let the buffer warm up before use - Urea-based Hybridization Buffer
Final concentration:
5x SSC
1% dextran
4 M urea
50 μg/ml of heparin
50 μg/ml of tRNA (or salmon sperm)
1% SDS
To prepare (10 ml) add in the following order, prepare freshly:
2.5 ml of 20x SSC
0.1 g of dextran powder (dissolve well)
5 ml of 8 M urea stock solution
25 μl of heparin (20 mg/ml stock)
10 μl tRNA
5.1 ml of MilliQ H2O
500 μl of 20% SDS
Notes:- Be aware that premature addition of SDS might cause a precipitate to appear.
- Do not agitate vigorously, to avoid bubbles.
- Hybridization buffer can also be aliquoted and stored at -20 °C, in this case avoid multiple thawing/freezing cycles.
- 50% Hybridization Buffer
50% Urea-based Hybridization Buffer
50% 1x PBST
Prepare freshly - Wash I
Final concentration:
4 M urea
0.1% Tween-20
5x SSC
Add MilliQ H2O to volume
Prepare freshly; or shortly beforehand (e.g., the evening before use) - Wash II
Final concentration:
2 M urea
0.1% Tween-20
2x SSC
Add MilliQ H2O to volume
Prepare freshly; or shortly beforehand (e.g., the evening before use) - Wash III
Final concentration:
0.1% Tween-20
2x SSC
Add MilliQ H2O to volume
Prepare freshly; or shortly beforehand (e.g., the evening before use) - 10x MAB buffer stock solution
- Add to 700 ml of MilliQ H2O: 116 g of maleic acid, 87 g of NaCl, 40 g of NaOH
- Adjust carefully the pH to 7.5, and top to 1 L with MilliQ H2O
- Store at room temperature, discard if precipitate/particles appear (the bottle might have been contaminated)
- 1x MABT buffer
- Dilute stock solution 1:10 with MilliQ H2O, add 0.1% Tween-20
- Store at room temperature, discard if precipitate/particles appear (the bottle might have been contaminated)
- 10x blocking reagent stock solution
- Dissolve in maleic acid buffer to a final concentration of 10% (w/v), shaking and heating to 60 °C
- Stock solution should be autoclaved, aliquoted and stored at -20 °C
- 1x Blocking buffer
- 5 M NaCl
- Dissolve 292 g of NaCl in 800 ml of MilliQ H2O
- Adjust the volume to 1 L with MilliQ H2O
- Autoclave for sterilizing
- Store at room temperature, discard if precipitate/particles appear (the bottle might have been contaminated)
- Dissolve 292 g of NaCl in 800 ml of MilliQ H2O
- 1 M Tris-HCl, pH 9.5
- Dissolve 121.1 g of Tris-base in 800 ml of MilliQ H2O
- Adjust the pH to 9.5 by adding HCl (let the solution reach room temperature for accurate adjustment of pH)
- Adjust the volume to 1 L with MilliQ H2O
- Autoclave for sterilizing
- Store at room temperature, discard if precipitate/particles appear (the bottle might have been contaminated)
- 1 M MgCl2
- Dissolve 203.3 g of MgCl2·6H2O in 800 ml of MilliQ H2O
- Adjust the volume to 1 L with MilliQ H2O
- Autoclave
- Store at room temperature, discard if precipitate/particles appear (the bottle might have been contaminated)
- NTMT buffer
Final concentration:
100 mM NaCl
100 mM Tris-HCl
50 mM of MgCl2
For 100 ml, add:
2 ml of 5 M NaCl
10 ml of 1 M Tris-HCl (pH 9.5)
5 ml of 1 M MgCl2
1 ml of Tween-20
MilliQ H2O to volume
Prepare freshly - NTMT minus buffer
Final concentration:
100 mM NaCl
100 mM Tris-HCl
For 100 ml add:
2 ml of 5 M NaCl
10 ml of 1 M Tris-HCl (pH 9.5)
1 ml of Tween-20
MilliQ H2O to volume
Prepare freshly - NBT/BCIP in NTMT buffer
- Add to NTMT buffer:
0.08 mg/ml of Nitro Blue Tetrazolium
0.1 mg/ml of 5-Bromo-4-Chloro-3-Indolyl-Phosphate - Prepare freshly and protect from light
- Add to NTMT buffer:
Acknowledgments
I thank Lucas Leclère and Evelyn Houliston for comments on the manuscript and discussions, and the other members of the Houliston/Momose group, in particular Sandra Chevalier, for discussion and assistance. I also thank all the people who contributed to testing this protocol on their organism of study, in particular Gonzalo Quiroga-Artigas, Lorenzo Ricci, Daniel Thiel. I thank the anonymous reviewers for their helpful comments on the manuscript. I also thank Enrique Arboleda for comments on the final version of the manuscript. Funding: Agence National de la Recherche, grant “MEDUSEVO” ANR-13-PDOC-0016.
Competing interests
The author declares there are no competing interests.
References
- Azaripour, A., Lagerweij, T., Scharfbillig, C., Jadczak, A. E., Willershausen, B. and Van Noorden, C. J. (2016). A survey of clearing techniques for 3D imaging of tissues with special reference to connective tissue. Prog Histochem Cytochem 51(2): 9-23.
- CICAD 31: N,N-Dimethylformamide, 2001. World Health Organization Geneva.
- Cox, K. H., DeLeon, D. V., Angerer, L. M. and Angerer, R. C. (1984). Detection of mRNAs in sea urchin embryos by in situ hybridization using asymmetric RNA probes. Dev Biol 101(2): 485-502.
- Eltoum, I., Fredenburgh, J., Myers, R. B. and Grizzle, W. E. (2001). Introduction to the theory and practice of fixation of tissues. Journal of Histotechnology 24(3): 173-190.
- Fail, P. A., George, J. D., Grizzle, T. B. and Heindel, J. J. (1998). Formamide and dimethylformamide: reproductive assessment by continuous breeding in mice. Reprod Toxicol 12(3): 317-332.
- Fontenete, S., Carvalho, D., Guimaraes, N., Madureira, P., Figueiredo, C., Wengel, J. and Azevedo, N. F. (2016). Application of locked nucleic acid-based probes in fluorescence in situ hybridization. Appl Microbiol Biotechnol 100(13): 5897-5906.
- George, J. D., Price, C. J., Marr, M. C., Myers, C. B. and Jahnke, G. D. (2000). Evaluation of the developmental toxicity of formamide in Sprague-Dawley (CD) rats. Toxicol Sci 57(2): 284-291.
- George, J. D., Price, C. J., Marr, M. C., Myers, C. B. and Jahnke, G. D. (2002). Evaluation of the developmental toxicity of formamide in New Zealand white rabbits. Toxicol Sci 69(1): 165-174.
- Gleich, J. (1974). Proceedings: The influence of simple acid amides on fetal development of mice. Naunyn Schmiedebergs Arch Pharmacol 282(Suppl): suppl 282:R225.
- Haimovich, G. and Gerst, J. E. (2018). Single-molecule fluorescence in situ hybridization (smFISH) for RNA detection in adherent animal cells. Bio-protocol 8(21): e3070.
- Herskovits, T. T. (1963). Nonaqueous solutions of DNA; denaturation by urea and its methyl derivatives. Biochemistry 2: 335-340.
- Herskovits, T. T. and Bowen, J. J. (1974). Solution studies of the nucleic acid bases and related model compounds. Solubility in aqueous urea and amide solutions. Biochemistry 13(27): 5474-5483.
- Huang, E., Talukder, S., Hughes, T. R., Curk, T., Zupan, B., Shaulsky, G. and Katoh-Kurasawa, M. (2011). BzpF is a CREB-like transcription factor that regulates spore maturation and stability in Dictyostelium. Dev Biol 358(1): 137-146.
- Hutton, J. R. (1977). Renaturation kinetics and thermal stability of DNA in aqueous solutions of formamide and urea. Nucleic Acids Res 4(10): 3537-3555.
- Kennedy, G. L. and Short, R. D. (1986). Biological effects of acetamide, formamide, and their monomethyl and dimethyl derivatives. Crit Rev Toxicol 17(2): 129-182.
- Lambert, D. and Draper, D. E. (2012). Denaturation of RNA secondary and tertiary structure by urea: simple unfolded state models and free energy parameters account for measured m-values. Biochemistry 51(44): 9014-9026.
- Leclère, L., Horin, C., Chevalier, S., Lapébie, P., Dru, P., Peron, S., Jager, M., Condamine, T., Pottin, K., Romano, S., Steger, J., Sinigaglia, C., Barreau, C., Quiroga Artigas, G., Ruggiero, A., Fourrage, C., Kraus, J. E. M., Poulain, J., Aury, J. M., Wincker, P., Queinnec, E., Technau, U., Manuel, M., Momose, T., Houliston, E. and Copley, R. R. (2019). The genome of the jellyfish Clytia hemisphaerica and the evolution of the cnidarian life-cycle. Nat Ecol Evol 3(5):801-810.
- Lim, W. K., Rosgen, J. and Englander, S. W. (2009). Urea, but not guanidinium, destabilizes proteins by forming hydrogen bonds to the peptide group. Proc Natl Acad Sci U S A 106(8): 2595-2600.
- Marmur, J. and Doty, P. (1961). Thermal renaturation of deoxyribonucleic acids. J Mol Biol 3: 585-594.
- Manil-Segalen, M., Luksza, M., Kanaan, J., Marthiens, V., Lane, S. I. R., Jones, K. T., Terret, M. E., Basto, R. and Verlhac, M. H. (2018). Chromosome structural anomalies due to aberrant spindle forces exerted at gene editing sites in meiosis. J Cell Biol 217(10): 3416-3430.
- Merkle, J. and Zeller, H. (1980). Studies on acetamides and formamides for embryotoxic and teratogenic activities in the rabbit (author's transl). Arzneimittelforschung 30(9): 1557-1562.
- Pardue, M. L. and Gall, J. G. (1969). Molecular hybridization of radioactive DNA to the DNA of cytological preparations. Proc Natl Acad Sci U S A 64(2): 600-604.
- Priyakumar, U. D., Hyeon, C., Thirumalai, D. and Mackerell, A. D., Jr. (2009). Urea destabilizes RNA by forming stacking interactions and multiple hydrogen bonds with nucleic acid bases. J Am Chem Soc 131(49): 17759-17761.
- Quiroga Artigas, G., Lapebie, P., Leclère, L., Takeda, N., Deguchi, R., Jekely, G., Momose, T. and Houliston, E. (2018). A gonad-expressed opsin mediates light-induced spawning in the jellyfish Clytia. Elife 7. e29555.
- Simard, C., Lemieux, R. and Cote, S. (2001). Urea substitutes toxic formamide as destabilizing agent in nucleic acid hybridizations with RNA probes. Electrophoresis 22(13): 2679-2683.
- Sinigaglia, C., Thiel, D., Hejnol, A., Houliston, E. and Leclère, L. (2018). A safer, urea-based in situ hybridization method improves detection of gene expression in diverse animal species. Dev Biol 434(1): 15-23.
- Sive, H. L., Grainger, R. M. and Harland, R. M. (2007). Baskets for in situ hybridization and immunohistochemistry. CSH Protoc 2007: pdb prot4777.
- Stula, E. F. and Krauss, W. C. (1977). Embryotoxicity in rats and rabbits from cutaneous application of amide-type solvents and substituted ureas. Toxicol Appl Pharmacol 41(1): 35-55.
- Thiel, D., Bauknecht, P., Jekely, G. and Hejnol, A. (2017). An ancient FMRFamide-related peptide-receptor pair induces defence behaviour in a brachiopod larva. Open Biol 7(8). pii: 170136.
- Tainaka, K., Kuno, A., Kubota, S. I., Murakami, T. and Ueda, H. R. (2016). Chemical principles in tissue clearing and staining protocols for whole-body cell profiling. Annu Rev Cell Dev Biol 32: 713-741.
Article Information
Copyright
© 2019 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Sinigaglia, C. (2019). A Widely Applicable Urea-based Fluorescent/Colorimetric mRNA in situ Hybridization Protocol. Bio-protocol 9(17): e3360. DOI: 10.21769/BioProtoc.3360.
Category
Developmental Biology > Morphogenesis > Organogenesis
Cell Biology > Cell imaging > Fixed-tissue imaging
Molecular Biology > RNA > RNA labeling
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









