- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Photoaffinity Labeling of Respiratory Complex I in Bovine Heart Submitochondrial Particles by Photoreactive [125I] amilorides
Published: Vol 9, Iss 17, Sep 5, 2019 DOI: 10.21769/BioProtoc.3349 Views: 4771
Reviewed by: Alexandros AlexandratosAlexander GalkinAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

In vitro Induction and Detection of Acrosomal Exocytosis in Human Spermatozoa
Shenae L. Cafe [...] Brett Nixon
Jul 20, 2020 4778 Views

A New Method for Studying RNA-binding Proteins on Specific RNAs
Weiping Sun [...] Min Zhuang
May 20, 2021 10582 Views

Flow Cytometry Analysis of Microglial Phenotypes in the Murine Brain During Aging and Disease
Jillian E. J. Cox [...] Sarah R. Ocañas
Jun 20, 2024 3568 Views
Abstract
The architecture of quinone/inhibitor-access channel in proton-translocating NADH-quinone oxidoreductase (respiratory complex I) was modeled by X-ray crystallography and cryo-EM, however, it remains debatable whether the channel model reflects the physiologically relevant state present throughout the catalytic cycle. Using photoreactive [125I]amilorides, we demonstrated that amiloride-type inhibitors bind to the interfacial region of multiple subunits (49-kDa, ND1, PSST, and 39-kDa subunits), which is difficult to reconcile with the current channel model. This report describes the procedures for photoaffinity labeling of bovine submitochondrial particles by photoreactive [125I]amilorides. The protocol could be widely applicable for the characterization of various biologically active compounds, whose target protein remains to be identified or characterized.
Keywords: Photoaffinity labelingBackground
Proton-translocating NADH-quinone oxidoreductase (respiratory complex I) is a multi-subunit membrane protein complex, which catalyzes the initial step of mitochondrial/bacterial electron transport chains (Hirst, 2013). The recent progress in X-ray crystallography and cryo-EM enabled modeling the entire structure of complex I. The structural studies proposed the quinone/inhibitor-access channel model to explain how quinone/inhibitor bind to the enzyme (Sazanov, 2015; Wirth et al., 2016). However, since there is currently no structural data of complex I with bound-quinone or inhibitor, it remains debatable whether the channel model reflects the physiologically relevant state present throughout the catalytic cycle.
Photoaffinity labeling technique, which uses a synthetic ligand that possesses photoreactive group such as diazirine and phenyl-azido, is the most commonly used method of affinity-based protein modification (Hatanaka and Sadakane, 2002). It provides a powerful means of investigating interactions between biologically active compounds and the proteins of interest. Recently, to get insights into the structure of quinone/inhibitor-binding site in mitochondrial complex I, we carried out photoaffinity labeling using photoreactive [125I]amilorides ([125I]PRA3, [125I]PRA4, [125I]PRA5, and [125I]PRA6) with bovine heart submitochondrial particles (SMPs) (Uno et al., 2019). The radioisotope (125I) was used as a tracer, which enables the experiment with the lowest concentrations of ligands (1-10 nM). These concentrations are approximately two orders of magnitude lower than those of alkyne-tagged amilorides (PRA1 and PRA2, Murai et al., 2015), which can be conjugated with biotin or fluorophores via Cu+-catalyzed click chemistry (Wang et al., 2003) after cross-linking reaction. This advantage may minimize the possibility of non-specific labeling, which is a primary cause of false-positive results.
We herein describe the detailed procedures of the photoaffinity labeling of complex I in bovine SMPs by [125I]PRA5 as an example (Figure 1), which includes 1) preparation of 125I-tagged photoreactive ligand, 2) cross-linking of bovine SMPs by the ligand, and 3) identification of the labeled protein(s).
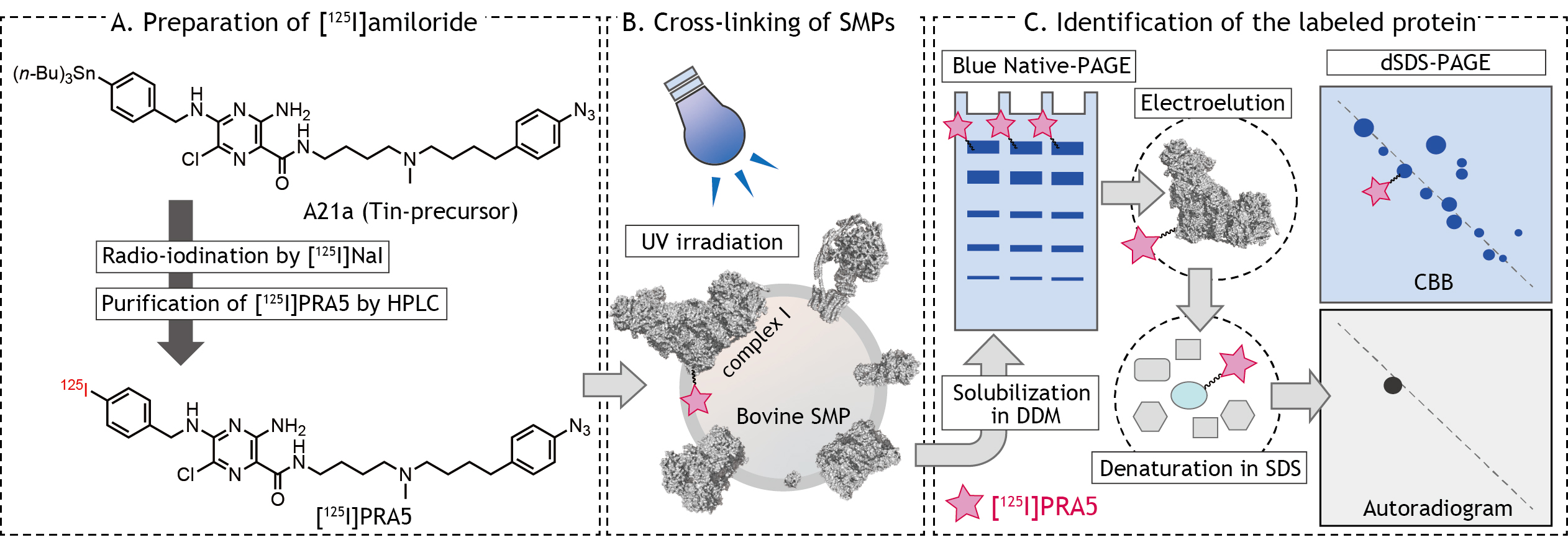
Figure 1. Schematic presentation of photoaffinity labeling of bovine heart SMPs by [125I]amilorides
Materials and Reagents
- 1.5 ml Screw-capped microtube (Watson, catalog number: 139-115-112C)
- 1.5 ml microtube (Safe-Lock Tube, Eppendorf, catalog number: 0030120086)
- Pipette tips (Gilson)
- Membrane filter unit (Amicon-Ultra, 100 kDa, Merck Millipore, catalog number: UFC510096)
- Reverse-phase HPLC column (COSMOSIL 5C18-MSII, 4.6 mm x 200 mm, Nacalai Tesque)
- TLC plate (Merck Millipore, catalog number: 1.05554.0001)
- Plastic wrap
- Amiloride-tin-precursor (A21a, 1.0 mM in ethanol, Figure 1A, The synthetic procedures are described in Uno et al., 2019)
- [125I]NaI (carrier-free, 2,000 Ci/mmol, Perkin-Elmer, catalog number: NEZ033A)
- Chloramine T (Wako Pure Chemicals, catalog number: 032-02182)
- Sodium hydrogen sulfite (NaHSO3, Wako Pure Chemicals, catalog number: 198-01371)
- Bovine submitochondrial particles (SMPs)
Notes:- SMPs are inside-out vesicles of inner mitochondrial membrane.
- Mitochondria were isolated from bovine heart as described elsewhere (Smith, 1967).
- SMPs were prepared on the basis of the previously described protocol (Matsuno-Yagi and Hatefi, 1985) using a sonication medium containing 0.25 M sucrose, 1.0 mM potassium succinate, 1.5 mM ATP, 10 mM MgCl2, 10 mM MnCl2, and 10 mM Tris/HCl (pH 7.4), followed by ultracentrifugation. They are stored in a buffer containing 250 mM sucrose and 10 mM Tris-HCl (pH 7.4) at -80 °C until used.
- NativePAGETM Bis-Tris protein gels (4-16% precast gel, Thermo Fisher, catalog number: BN1004BOX)
- NativePAGETM Runnning buffer and Cathode buffer additive (Thermo Fisher, catalog numbers: BN2001 and BN2002, respectively)
- Serva Blue G (CBB G-250, Serva, catalog number: 35050.01)
- n-Dodecyl-D-β-maltoside (DDM, Dojin, catalog number: D316)
- Glycerol (Wako Pure Chemicals, catalog number: 075-00616)
- Ethanol (Wako Pure Chemicals, catalog number: 057-00456)
- Methanol (Wako Pure Chemicals, catalog number: 137-01823)
- Acetic acid (Wako Pure Chemicals, catalog number: 017-00251)
- Sucrose (Wako Pure Chemicals, catalog number: 192-00017)
- MgCl2 (hexahydrate, Wako Pure Chemicals, catalog number: 135-00165)
- NaCl (Wako Pure Chemicals, catalog number: 195-01665)
- Ponceau S (Nacalai Tesque, catalog number: 28322-72)
- Tricine (Dojin, catalog number: GB05)
- SDS (Wako Pure Chemicals, catalog number: 191-07145)
- Mercaptethanol (Wako Pure Chemicals, catalog number: 137-06862)
- Tris (Wako Pure Chemicals, catalog number: 011-20095)
- Acrylamide (Wako Pure Chemicals, catalog number: 011-08015)
- Bis-acrylamide (Wako Pure Chemicals, catalog number: 134-15081)
- Urea (Wako Pure Chemicals, catalog number: 219-00175)
- Silver Stain Kit (Wako Pure Chemicals, catalog number: 291-50301, compatible with MS)
- Trifluoroacetic acid (TFA)
- NADH (Oriental Yeast, catalog number: 44320000)
- Labeling buffer (see Recipes)
- 4x BN-PAGE sample buffer (see Recipes)
- Elution buffer (see Recipes)
- 4x Schägger sample buffer (see Recipes)
- 10x cathode buffer (see Recipes)
- 10x anode buffer (see Recipes)
- AB-3 mix (see Recipes)
- 3x gel buffer (see Recipes)
- 1.0 M KPi buffer, pH 7.4 (see Recipes)
- Gel composition for dSDS-PAGE (see Recipes)
Equipment
- Pippetmann (Gilson)
- -80 °C freezer (PHC, model: MDF-DU300H)
- HPLC system (Shimazdu, model: LC-10AS)
- Micro-centrifuge (CHIBITAN, Merck Millipore, model: XX42CF0RT)
- γ-counter (COBRATM II, Packard)
- Vacuum centrifugal evaporator (EYELA, model: CVE-2200)
- Bio-imaging analyzer (FLA-5100 or Typhoon FLA-9000 , Fuji Film or GE healthcare, respectively)
- Imaging plate (BAS IP MS 2040E, GE Healthcare)
- Vortex mixer (AS ONE, model: Trio TM-1N)
- Heat block (AS ONE, model: MyBL-10)
- Long wavelength UV lamp (UVP, Black-lay model B-100AP, equipped with a 100 W bulb, 365 nm)
- Electro-Eluter (Bio-Rad, model: 422)
- Gel dryer (Bio-Rad, Model583, equipped with HydroTech vacuum pump)
- Gel electrophoresis apparatus and glass plates (ATTO, AE-6500, 8 x 9 cm for mini-size gel format)
Software
- Image analysis software such as Multi Gauge or Image Quant (Fuji Film or GE Healthcare, respectively)
Procedure
- Preparation of photoreactive [125I]amilorides ([125I]PRA5 as an example)
- Prepare ethanolic solution of Tin-precursor A21a (1.0 mM, 20 µl) in a screw-capped 1.5 ml microtube.
- Add [125I]NaI (1 mCi, 2,000 Ci/mmol, 10 μl).
Note: [125I]NaI should be handled in a draft chamber until the reaction is quenched by NaHSO3. - Initiate the reaction by the addition of aqueous chloramine T (3.0 mM in 1.0 M KPi buffer, pH 7.4, 10 µl).
Note: Aqueous chloramine T solution should be freshly prepared for each experiment. - Vortex and spin-down the tube.
- Incubate the mixture for 10 min at room temperature.
- Quench the reaction by the addition of 5% (w/v) aqueous sodium hydrogen sulfite (10 µl).
- Inject whole volume of the mixture (~50 µl) to the HPLC with a manual injection port (loop volume should be larger than 100 µl). LC conditions are as follows:
- Column: COSMOSIL 5C18-MSII, 4.6 mm x 200 mm (Nacalai Tesque)
- Solvent: methanol/0.01% aqueous TFA (step gradient elution of 1:9, then 9:1).
- Temperature: 35 °C
- Flow rate: 0.8 ml/min.
- Collect fractions every 30 s (~400 µl).
- Take 3 µl of the samples to assess the radioactivity and purity of each fractions by γ-counter (2 µl) and radio-TLC (1 µl), respectively (see Figure 2).
- Combine the fractions containing [125I]PRA5 (frs. 8 and 9 in Figure 2), and concentrate the sample up to ~50 µl by a vacuum-centrifugal evaporator.
Note: Do not dry the sample completely. - Adjust the concentration of [125I]PRA5 to 1 mCi/ml by ethanol.
- Store the sample at 4 °C ([125I]amilorides are stable as ethanolic solutions for at least 2 months).
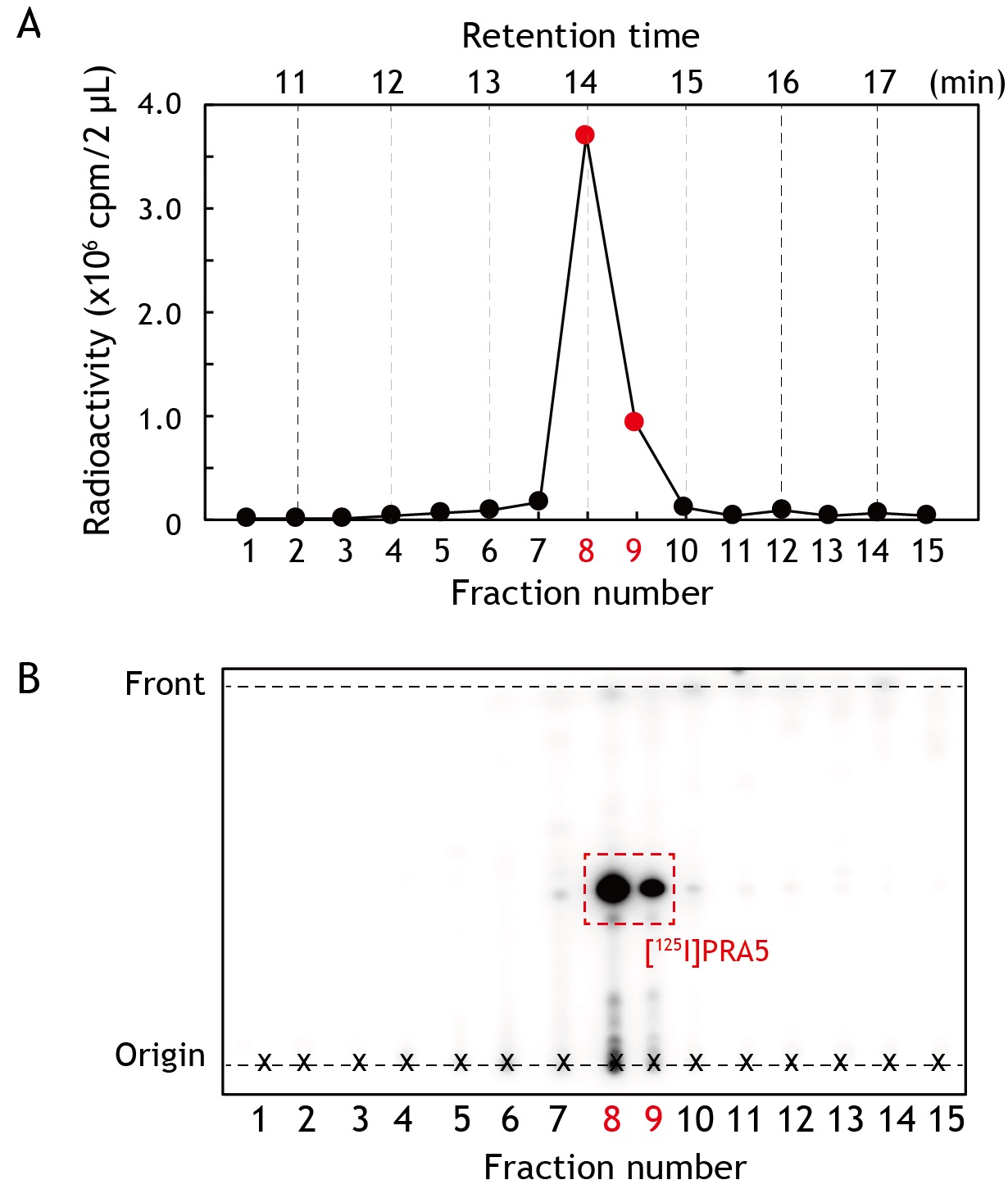
Figure 2. Purification of photoreactive [125I]PRA5 by reverse phase HPLC. The mixture containing crude [125I]PRA5 was applied to a C18 column. The salts were removed by rinsing the column with 10% methanol/0.01% aqueous TFA (0-10 min), then [125I]PRA5 was eluted with an isocratic 90% methanol/0.01% aqueous TFA (10-20 min). The elute was fractionated every 30 s, and 2 µl aliquot from each fraction was transferred to RIA tube and the radioactivity was measured using γ-counter (retention time; 10-17.5 min). A. Radioactivity distribution in HPLC chromatogram. The radiochemical yield was approximately 70% from the initial [125I]NaI. B. TLC analysis of the collected fractions. The samples (1 µl) were analyzed on a TLC plate using 10% methanol/chloroform as a mobile phase. The plate was exposed on an imaging plate for 1 h and analyzed by Bio-imaging analyzed FLA-5100.
- Cross-linking of bovine SMPs by [125I]PRA5
- In a 1.5 ml microtube, add the suspension of bovine SMPs in the labeling buffer (2.0-4.0 mg/ml, 100-200 µl).
- Add [125I]PRA5 at the concentration of 5-10 nM.
Note: Ethanol concentration should be below ~3%. - Incubate at room temperature for 10 min.
- Add 50 µM NADH, then incubate for 5 min.
- Irradiate with a long wavelength UV lamp on ice for 10 min at the distance of 10 cm from the light source (Figure 3).
- Quench the cross-linking reaction by adding 10% (w/v) DDM to a final concentration 1.0%.
- Incubate the mixture on ice for 1 h.
- Add 4x BN-PAGE sample buffer.

Figure 3. A set-up for photoaffinity labeling of bovine SMPs. The 1.5 ml Eppendorf tubes containing SMPs are placed with open lids on ice. They are UV-irradiated for 10 min at the distance of approximately 10 cm from the light source. For efficient labeling, the volume of the sample should be 100-200 µl/tube.
- Identification of the protein labeled by [125I]PRA5
- Isolation of complex I by Blue Native (BN)-PAGE and electroelution (Schägger, 2006)
- Add 5% Serva Blue G (CBB G-250). The CBB/DDM ratio should be adjusted to be 1/4 by weight.
- Load samples into the 4-16% BN-precast gel (100-200 µg protein/well).
- Start electrophoresis (100 V, 15 mA, for 1h).
- Increase the voltage to 200 V (15 mA), and continue electrophoresis until the front dye reaches the bottom of the gel.
- Excise the complex I band with a razor blade.
Note: Do not fix the BN gel. Do not include any unstained (or poorly stained) gel. Excise only the heart of the complex I band. - Assemble the Electro-Eluter according to the manufacturer’s protocols.
- Fill the glass tube and the Eluter with elution buffer, and place the gel piece in the tube (4-5 gel pieces/tube).
- Elute protein at 10 mA/glass tube for overnight in a cold room.
Note: If the elution is successful, the gel piece becomes colorless. - Collect the eluted protein according to the manufacturer’s protocol (We recommend washing the dialysis membrane with 50-100 µl of 0.5% SDS to solubilize the precipitated proteins).
- Concentrate the collected solution with Amicon-Ultra 0.5 (100 kDa cut-off) to the appropriate volume (50-100 µl).
- Store the sample at -80 °C until used.
- Resolution of complex I subunits by doubled SDS-PAGE (dSDS-PAGE, Rais et al., 2004; Wittig, 2006)
- Add 4x Schägger sample buffer to the complex I isolated by electroelution.
- Incubate the mixture at 40 °C for 1 h.
Note: Do not boil the samples because some hydrophobic subunits in complex I aggregate. - Separate complex I proteins on a first dimensional Schägger-type SDS gel (10% T, 3% C containing 6.0 M urea with 4% stacking gel, Table 1) with a voltage of 30 V (40 mA).
- When the samples have completely entered the gel, increase the voltage to 100 V, and continue electrophoresis until the dye front reaches the bottom of the gel.
- Excise a first dimensional gel strip and incubate it for 30 min in an acidic buffer containing 100 mM Tris/HCl (pH 2.0), then fix the strip between two glass plates (see Figures 4A, 4B, and 4C).
- Pour the second dimensional acrylamide mixture (16% T, 3% C, Table 1) and overlay with water (see Figure 4D).
- After polymerization, push down the first dimensional gel strip and make the two gels stick together (see Figure 4E).
- Fill the gap with acrylamide mixture (16% T, 3% C, Table 1) containing 150 mM Tris/HCl (pH 7.4) and trace of Serva Blue G (spacer gel, see Figure 4F).
- Start second dimensional electrophoresis with a voltage of 30 V (40 mA).
- When the samples have completely entered the second dimensional gel, the voltage can be increased to 150 V.
- Silver stain and autoradiography
- Fix and stain the gel with silver by an appropriate method (Yan et al., 2000) or by commercially available silver stain kit.
- Scan the silver-stained gel using a conventional flatbed scanner.
- Before place the gel on the gel drier, equilibrate the gel in a solution containing 3% glycerol, 40% methanol, and 10% acetic acid for 30 min.
- Dry the gel using gel drier at 65 °C for 3 h. Temperature should be raised slowly [about 1 °C/min (“Gradient mode”)].
- When the gel is completely dried, cover the gel with plastic wrap, then expose it onto the imaging plate for 12-24 h.
- Analyze the migration pattern of radio-labeled proteins by Bio-imaging analyzer, and compare those of silver-stained protein (representative results are shown in Figure 5).
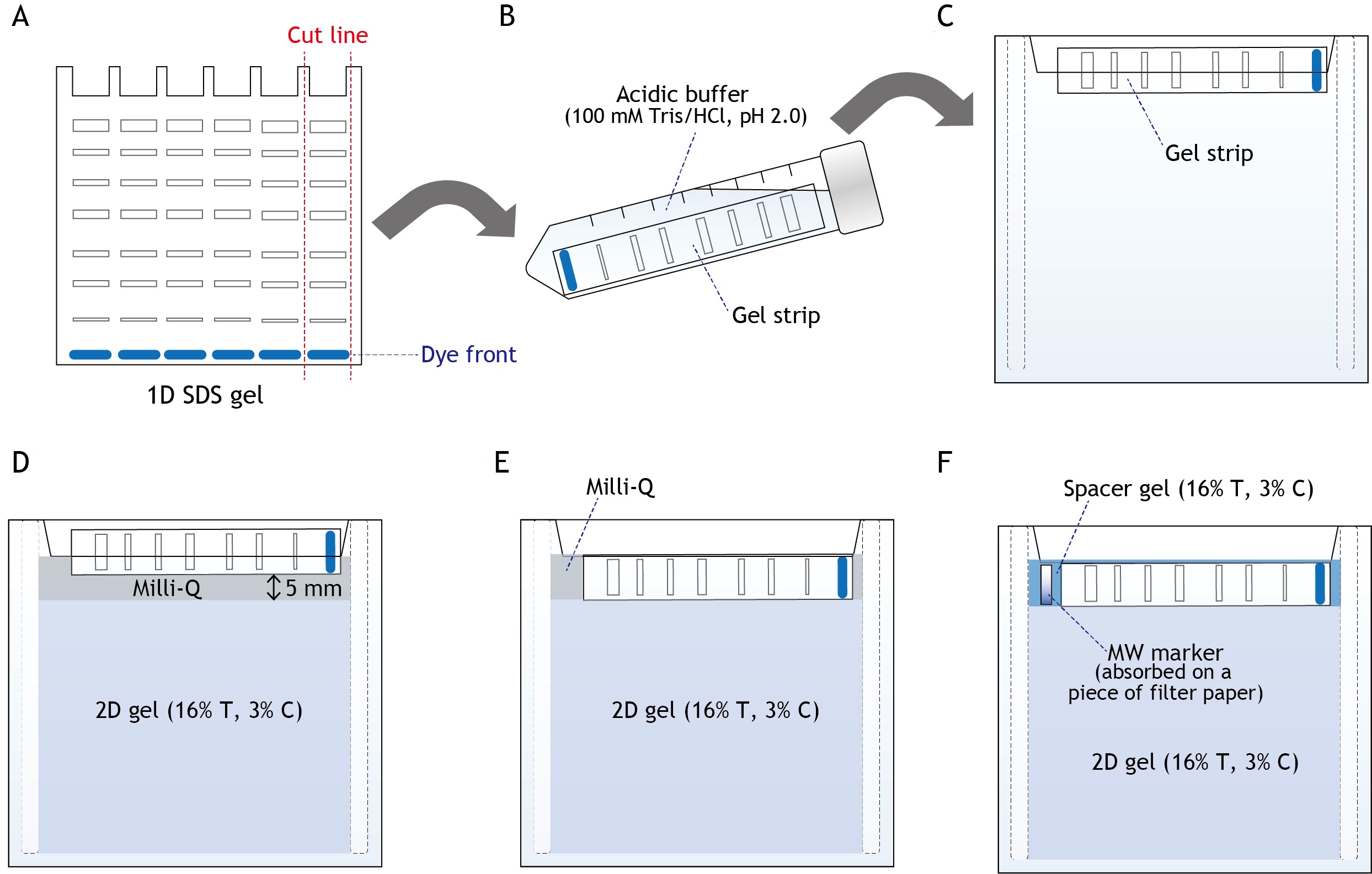
Figure 4. Preparation of the second dimensional gel for dSDS-PAGE. A. Excise the first dimensional gel strip. B. Carefully place the gel strip to a 15 ml falcon tube containing an acidic buffer. Incubate the gel for 30 min at room temperature. C. Fix the strip between two glass plates. D. Pour the second dimensional acrylamide mixture, and overlay with Milli-Q H2O. E. After polymerization of the acrylamide gel mixture, push down the gel strip to make the two gels stick together. F. Remove Milli-Q, then fill the gap with a spacer gel.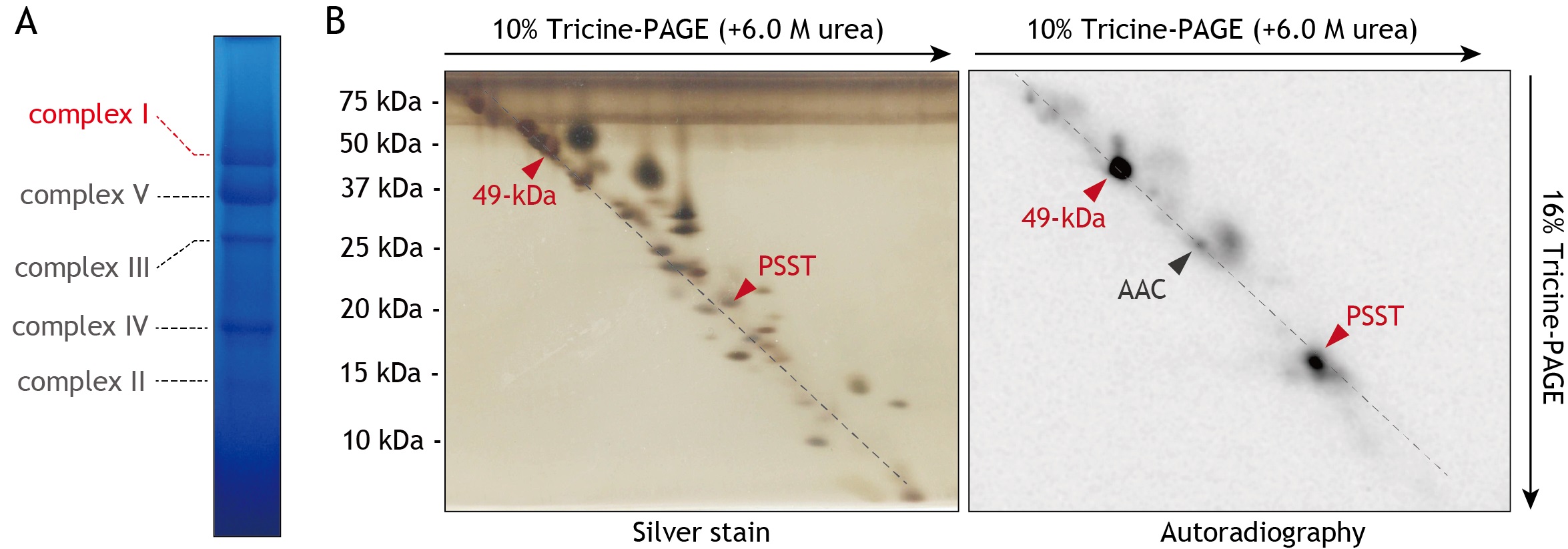
Figure 5. Analysis of the proteins in SMPs labeled by [125I]PRA5. A. Separation of complex I by BN-PAGE. The SMPs labeled by [125I]PRA5 were separated on a 4-16% BN gel. The complex I band was identified by activity stain by NADH/NBT system, as described elsewhere (Murai et al., 2009; Shiraishi et al., 2012), and subjected to electroelution. Approximately 100 µg of SMPs proteins were loaded into each well. B. Resolution of complex I by dSDS-PAGE. The [125I]PRA5-labeled complex I, purified by BN-PAGE and electroelution, was separated on a first dimensional 10% Schägger-type Tricine gel (10% T, 3% C containing 6.0 M urea), followed by second dimensional separation on a 16% Schägger-type gel (16% T, 3% C). The 2D gel was subjected to silver stain (left) and autoradiography (right). The labeling by [125I]PRA5 provided two radioactive spots corresponding to the 49-kDa and PSST subunits, both of which comprise the quinone/inhibitor-binding pocket of complex I (Hirst 2013 and Sazanov 2015). The spots were identified by mass spectrometry and Western blotting (Shiraishi et al., 2012). We note that the weak radioactivity was found in a protein spot of ADP/ATP carrier (AAC), which was co-purified with complex I. The gel images are the same as those used in Figure 7C of the reference (Uno et al., 2019).
- Isolation of complex I by Blue Native (BN)-PAGE and electroelution (Schägger, 2006)
Recipes
- Labeling buffer (for UV irradiation of bovine SMP)
250 mM sucrose
50 mM KPi (pH 7.4)
1 mM MgCl2 - 4x BN-PAGE sample buffer
200 mM Bis-Tris/HCl (pH 7.2)
200 mM NaCl
40% (w/v) glycerol
0.004% (w/v) Ponceau S - Elution buffer (for electroelution of complex I)
25 mM Tricine
7.5 mM Bis-Tris/HCl (pH 7.0 adjusted at 4 °C) - 4x Schägger sample buffer (for Tricine-PAGE)
150 mM Tris/HCl (pH 7.0)
12% (w/v) SDS
30% (w/v) glycerol
6% (w/v) mercaptoethanol
0.05% (w/v) Serva Blue G - 10x cathode buffer (for Schägger-type SDS-PAGE)
1.0 M Tris
1.0 M Tricine
1% SDS - 10x anode buffer (for Schägger-type SDS-PAGE)
1.0 M Tris/HCl (pH 8.9) - AB-3 mix (49.5% T, 3% C, for Schägger-type SDS-PAGE)
48% acrylamide
1.5% bis-acrylamide - 3x gel buffer (for Schägger-type SDS-PAGE)
3.0 M Tris/HCl (pH 8.45)
0.3% SDS - 1.0 M KPi buffer, pH 7.4
Dissolve K2HPO4 (12.1 g, 69.5 mmol) and KH2PO4 (4.1 g, 30.4 mmol) in 100 mL of distilled water. - Gel composition for dSDS-PAGE (for Schägger-type SDS-PAGE) (Table 1)
Table 1. Preparation of Schägger-type SDS gels (for two ATTO mini-size gels)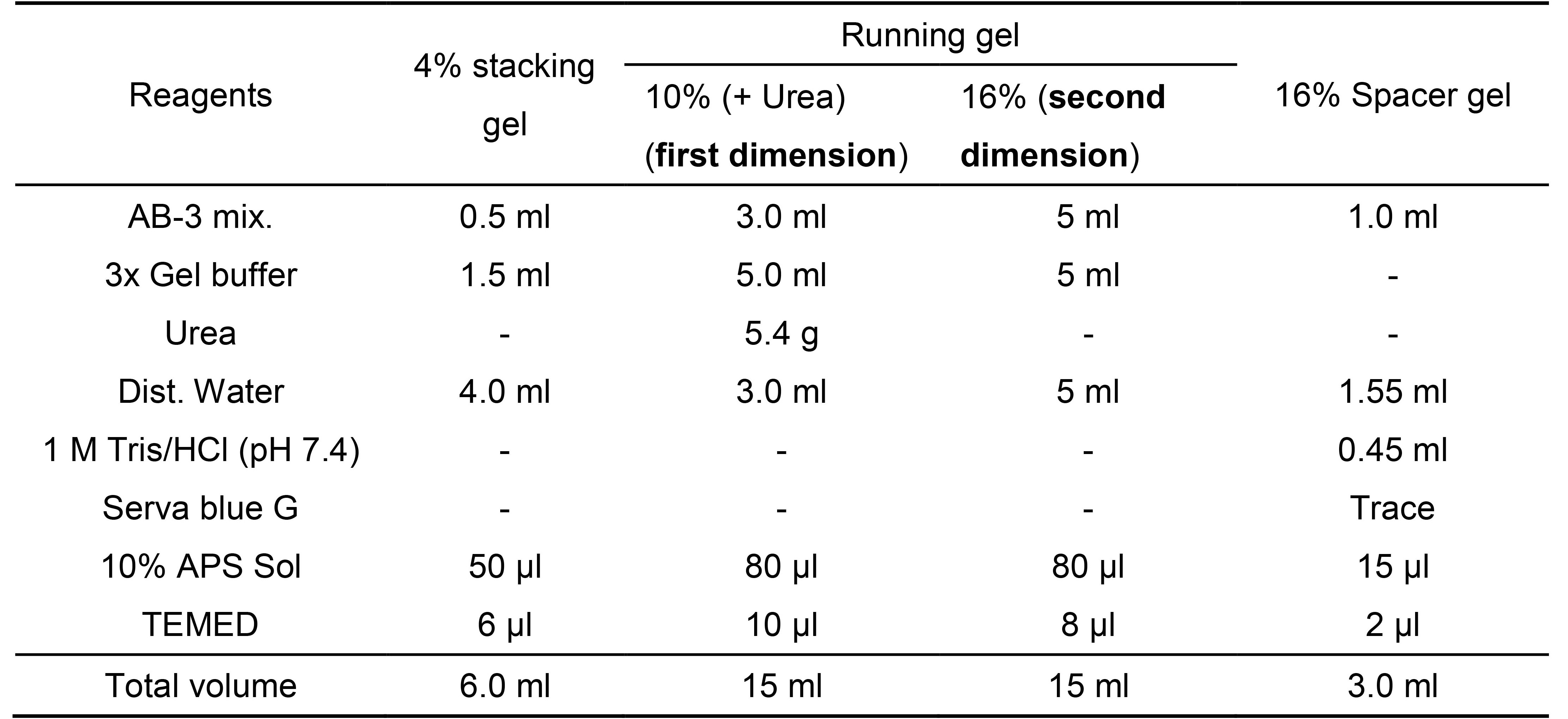
Acknowledgments
AcknowledgmentsThis protocol was adapted from Uno et al. (2019). This study was supported by JSPS KAKENHI (Grant Numbers JP26292060 and JP18H02147 to H.M., Grant Number JP18K05458 to M.M.). The experiments involving radioisotope techniques were performed at the Radioisotope Research Center, Kyoto University.
Competing interests
The authors declare that they have no conflicts of interest with the contents of this article.
References
- Hatanaka, Y. and Sadakane, Y. (2002). Photoaffinity labeling in drug discovery and developments: chemical gateway for entering proteomic frontier. Curr Top Med Chem 2(3): 271-288.
- Hirst, J. (2013). Mitochondrial complex I. Annu Rev Biochem 82: 551-575.
- Matsuno-Yagi, A. and Hatefi, Y. (1985). Studies on the mechanism of oxidative phosphorylation. Catalytic site cooperativity in ATP synthesis. J Biol Chem 260(27): 11424-11427.
- Murai, M., Sekiguchi, K., Nishioka, T. and Miyoshi, H. (2009). Characterization of the inhibitor binding site in mitochondrial NADH-ubiquinone oxidoreductase by photoaffinity labeling using a quinazoline-type inhibitor. Biochemistry 48(4): 688-698.
- Murai, M., Murakami, S., Ito, T. and Miyoshi, H. (2015). Amilorides bind to the quinone binding pocket of bovine mitochondrial complex I. Biochemistry 54(17): 2739-2746.
- Rais, I., Karas, M. and Schägger, H. (2004). Two-dimensional electrophoresis for the isolation of integral membrane proteins and mass spectrometric identification. Proteomics 4(9): 2567-2571.
- Sazanov, L. A. (2015). A giant molecular proton pump: structure and mechanism of respiratory complex I. Nat Rev Mol Cell Biol 16(6): 375-388.
- Schägger, H. (2006). Tricine-SDS-PAGE. Nat Protoc 1(1): 16-22.
- Shiraishi, Y., Murai, M., Sakiyama, N., Ifuku, K. and Miyoshi, H. (2012). Fenpyroximate binds to the interface between PSST and 49 kDa subunits in mitochondrial NADH-ubiquinone oxidoreductase. Biochemistry 51(9): 1953-1963.
- Smith, A. L. (1967). Mitochondria : Slaughterhouse Material, Small-Scale. Methods Enzymol 10: 81-86.
- Uno, S., Kimura, H., Murai, M. and Miyoshi, H. (2019). Exploring the quinone/inhibitor-binding pocket in mitochondrial respiratory complex I by chemical biology approaches. J Biol Chem 294(2): 679-696.
- Wang, Q., Chan, T. R., Hilgraf, R., Fokin, V. V., Sharpless, K. B. and Finn, M. G. (2003). Bioconjugation by copper(I)-catalyzed azide-alkyne [3 + 2] cycloaddition. J Am Chem Soc 125(11): 3192-3193.
- Wirth, C., Brandt, U., Hunte, C. and Zickermann, V. (2016). Structure and function of mitochondrial complex I. Biochim Biophys Acta 1857(7): 902-914.
- Wittig, I., Braun, H. P. and Schägger, H. (2006). Blue native PAGE. Nat Protoc 1(1): 418-428.
- Yan, J. X., Wait, R., Berkelman, T., Harry, R. A., Westbrook, J. A., Wheeler, C. H. and Dunn, M. J. (2000). A modified silver staining protocol for visualization of proteins compatible with matrix‐assisted laser desorption/ionization and electrospray ionization‐mass spectrometry. Electrophoresis 21(17): 3666-3672.
Article Information
Copyright
© 2019 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Murai, M. and Miyoshi, H. (2019). Photoaffinity Labeling of Respiratory Complex I in Bovine Heart Submitochondrial Particles by Photoreactive [125I] amilorides. Bio-protocol 9(17): e3349. DOI: 10.21769/BioProtoc.3349.
- Uno, S., Kimura, H., Murai, M. and Miyoshi, H. (2019). Exploring the quinone/inhibitor-binding pocket in mitochondrial respiratory complex I by chemical biology approaches. J Biol Chem 294(2): 679-696.
Category
Biochemistry > Other compound > Small molecule drugs
Biochemistry > Protein > Labeling
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









