- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Isolation of Powdery Mildew Haustoria from Infected Barley
Published: Vol 9, Iss 14, Jul 20, 2019 DOI: 10.21769/BioProtoc.3299 Views: 7831
Reviewed by: Amey RedkarThomas TorodeHailong Guo

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Quantitative Estimation of Auxin, Siderophore, and Hydrogen Cyanide Production in Halo and Drought-Tolerant Bacterial Isolates for Cucumber Growth
Zeinab Fotoohiyan and Ali Salehi Sardoei
Oct 5, 2025 1342 Views

A Reliable In Planta Inoculation and Antifungal Screening Protocol for Rhizoctonia solani-Induced Sheath Blight in Rice
Alinaj Yasin [...] Palash Deb Nath
Nov 5, 2025 1568 Views

Reproducible Emu-Based Workflow for High-Fidelity Soil and Plant Microbiome Profiling on HPC Clusters
Henrique M. Dias [...] Christopher Graham
Jan 20, 2026 404 Views
Abstract
Blumeria graminis is a fungus that causes powdery mildews on grasses, such as barley. Investigations of this pathogen present many challenges due to its obligate biotrophic nature. This means that the fungus can only grow in the presence of a living host plant. B. graminis forms epiphytic mycelia on the plant surface and feeding organs (haustoria) inside the epidermal cells of the host plant. Therefore, it is difficult to separate the fungus from plant tissues. This protocol shows how to obtain different fungal structures from powdery mildew infected barley leaves. The epiphytic mycelia including conidia and conidiophores can be separated after immersing the infected leaves into 5% cellulose acetate dissolved in acetone, and peeling off the cellulose acetate membrane. Then, the haustoria are isolated from dissected epidermis after cellulase degradation of plant cell walls. The isolated haustoria remain intact with few plant impurities. The haustoria may be visualized by epifluorescence microscopy after staining with the chitin-specific dye WGA-Alexa Fluor 488. Finally, dissected material can be either processed immediately or kept at -80 °C for long-term storage for studies on gene expression and protein identification, for example by mass spectrometry.
Keywords: Powdery mildewBackground
Powdery mildew of cereals, caused by Blumeria graminis, leads to significant yield loss (Spanu et al., 2010). The barley powdery mildew fungus B. graminis f. sp. hordei, is economically important, and one of the best studied powdery mildews (Both et al., 2005a; Bindschedler et al., 2009). This fungus is an obligate biotroph, which means it can only infect and propagate on living barley plants (Pedersen et al., 2012). Its asexual life cycle involves an intimate relationship with the host plant. Airborne conidia germinate on the host producing first a primary and then a secondary germ tube. The secondary germ tube develops an appressorium, from which a hyphal peg penetrates into an epidermal cell. Inside the cell, a penetration peg enlarges and forms a multidigitate feeding structure: the haustorium. This is surrounded by a plant-derived extrahaustorial membrane (Both et al., 2005b; Bindschedler et al., 2016).
Several studies of B. graminis f. sp. hordei have characterized gene expression profiles and proteomics of this fungus (Bindschedler et al., 2011; Pennington et al., 2016). However, the obligate biotrophic nature of B. graminis poses exceptional challenges to the investigations of this fungus, especially the studies of haustoria. The intracellular haustorium and the extrahaustorial membrane constitute a special compartment, which is functionally essential for the interaction with the host (Wang et al., 2009; Pliego et al., 2013). Investigating haustoria is critical for further understanding the mechanism of nutrient uptake into the pathogen as well as plant-pathogen recognition. Therefore, obtaining purified haustoria from infected plants has the potential of contributing significantly to these studies.
Previously, the procedure for isolating haustoria from obligate biotrophs employed affinity chromatography on rust fungi (Catanzariti et al., 2006; Garnica et al., 2014). During the 1990s, several centrifugation-based methods were introduced to isolate powdery mildew haustoria from pea plants (Mackie et al., 1991; Testut et al., 1999). This method was then applied to powdery mildews of other plants, including Arabidopsis (Wang et al., 2009; Micali et al., 2011). More recently, a filtration and gradient centrifugation-based method was described to isolate powdery mildew haustoria from barley (Godfrey et al., 2009). The hallmark of these published methods is that they are relatively laborious and require several different kinds of isolation buffers, different sizes of steel meshes and multiple centrifugation steps. Besides, the isolation procedure was required to be performed on ice. In this protocol, we developed a simpler way to isolate B. graminis haustoria from infected barley leaves. Releasing of haustoria from plant cells is achieved by enzymatically degrading the epidermal cell walls, and the purification of haustoria by filtration through a nylon mesh. This method requires digestion of the host plant cell walls, a single filtration and one centrifugation step.
Materials and Reagents
- Pipette tips
- Dedicated enclosed transparent container
- Razor blade
- Parafilm
- Pasteur pipettes
- 15 ml/50 ml polypropylene tubes
- Liquid nitrogen container
- Microscope glass slide
- Cover glass
- Barley (Hordeum vulgare) cv. Golden Promise seedlings, barley seeds were sown in John Innes 1 compost mixed with vermiculite (4:1) in 12 x 12 cm pots
- Liquid nitrogen
- Cellulose acetate
- Anhydrous acetone
- MES hydrate powder
- HCl
- WGA, Alexa Fluor 488 conjugate (Thermo Fisher Scientific, catalog number: W11261)
Note: This dye exhibits the bright, green fluorescence of the Alexa Fluor® 488 dye (excitation/emission maxima ~495/519 nm). - Cellulase Onozuka R-10 (Duchefa Biochemie, catalog number: C8001)
Note: “Cellulase Onozuka R-10” from Trichoderma viride.1 unit (U) of Cellulase will release 1.0 μmole of glucose from carboxymethyl cellulose. - NaCl
- KCl
- Na2HPO4
- KH2PO4
- PBS buffer (see Recipe 1)
Equipment
- Forceps
- Pipettes
- Scissor
- Tweezers
- 70 μm nylon mesh sieve
- Incubator
- Centrifuge
- GFP filter
- Epifluorescence microscope
- -80 °C freezer
Procedure
- Dissection of infected barley leaves
- Grow barley (Hordeum vulgare) cv. Golden Promise seedlings in a 16-h light/8-h dark cycle at 20 °C. It is essential to keep the plants well-watered, as drought stress prevents effective dissection of the epidermis.
- Maintain the powdery mildew fungus (B. graminis f. sp. hordei) on living barley plants in a dedicated enclosed transparent container protected from air currents that may displace the conidia. Remove old conidia by shaking the inoculum pots one day before infecting healthy plants.
- Inoculate seven-day barley seedlings. At this stage, the primary leaves are fully extended. The inoculation B. graminis is carried out by shaking heavily infected plants over the uninfected barley. The plants will be ready for dissection seven days after inoculation.
Note: It is highly recommended that a tight-fitting face mask be used when carrying out this procedure to prevent conidia from entering the operator’s airways. - Prepare scissor, tweezers, razor blade, Parafilm, Pasteur pipettes, 15 ml/50 ml polypropylene tubes and liquid nitrogen. Place two 50 ml tubes in a liquid nitrogen container and fill with liquid nitrogen.
- Dissolve 5 g of cellulose acetate in 100 ml anhydrous acetone. Pour the solution into a 15 ml polypropylene tube.
- Dissolve 0.2 g of MES hydrate powder in 100 ml H2O to make 10 mM MES buffer. Adjust the pH to 5.3 with HCl. Pour 20 ml of MES buffer into a 50 ml tube at room temperature.
- Cut one infected primary leaf and immerse it in 5% cellulose acetate acetone, holding it at the leaf base. Carefully take out the leaf and wait for a few seconds above the mouth of the tube to remove any cellulose acetate acetone droplets accumulating at the leaf tip. Then place the leaf onto two Pasteur pipettes.
- Repeat the above step five times. Let the acetone evaporate from the leaf surface for 5-8 min.
Note: This step is critical: the cellulose acetate membrane needs to be dry enough to allow peeling; however, if the leaves are left too long they dehydrate and wilt rendering the dissection of the epidermis impossible. - As the acetone evaporates, the epiphytic mycelia-containing cellulose acetate layer will harden and begin to peel away from the rest of the leaf. Now peel off the cellulose acetate membrane with the epiphytic fungal structures embedded in it. Keep the leaves stripped of epiphytic structures on water for 5 min to ensure full hydration.
- Following this, turn the leaf so the adaxial surface faces upwards. Press a razor gently near the tip of the leaf, carefully rock the blade back and forward.
Note: This step is challenging and requires some trial and error to gain sufficient experience to achieve correct dissection. The aim is to cut through the upper, adaxial epidermis and part of the mesophyll, while retaining the lower, abaxial epidermis intact. - Turn the leaf over to let the abaxial side facing upwards. While gripping the leaf at its tip (this forms a ‘tab’ which can be used to pull the epidermis) and holding the section of the leaf below the cut steady, pull back the abaxial epidermis which contains fungal haustoria.
Note: Free the epidermal layer by pushing away other layers with forceps, if needed. - Once free, carefully peel the abaxial epidermis in a continuous smooth motion.
- Once complete, take away the tip of the leaf (the ‘tab’) by cutting the epidermis and removing it with the tweezers.
Note: To isolate haustoria from epidermis, skip to Step B1. - Snap freeze peeled epiphytic and epidermal materials separately in two 50 ml polypropylene tubes in liquid nitrogen for future use.
- Store at -80 °C till needed.
- Grow barley (Hordeum vulgare) cv. Golden Promise seedlings in a 16-h light/8-h dark cycle at 20 °C. It is essential to keep the plants well-watered, as drought stress prevents effective dissection of the epidermis.
- Haustoria isolation
- Float peeled epidermis in 15 ml 10 mM MES hydrate buffer (pH 5.3) in a 50 ml tube.
- Once sufficient epidermis strips are collected (we used one pot in Video 1), add 200 mg of Cellulase R-10 into 10 ml MES buffer in a new 50 ml tube. Mix thoroughly, so the final concentration of cellulase is 2% (w/v).
- Carefully transfer all dissected epidermis with tweezers into the new mixture.
- Incubate the suspension at 28 °C for two hours with gentle shaking (e.g., 80 rpm on a rotating platform). During incubation, the cellulase will lyse the barley epidermal cells and release haustoria into the incubation buffer.
- After incubation, place a 70 μm nylon mesh sieve over a 50 ml tube. Slowly pour the buffer through the filter while using forceps as a barrier to retain larger pieces of undigested plant material.
- Once all the buffers are collected, centrifuge in a swing out rotor at 3,270 x g for 10 min, at 4 °C.
- After centrifugation, the pellet should contain haustoria.
- Pour off the supernatant and retain the pelleted haustoria.
Note: After this step, the haustoria can either be kept at -80 °C for long-term storage or stained immediately to visualize (continue with Procedure C).
- WGA-Alexa Fluor 488 staining (optional for quality control)
- Gently resuspend the haustorial pellet in 300 μl of PBS buffer (pH 7.4) (Recipe 1).
- Add WGA-Alexa Fluor 488 into the mixture to a final concentration of 10 μg/ml.
- Mix well by inverting the tube several times.
- Load the mixture onto a microscope glass slide and carefully place a cover glass on top.
- Visualize under the GFP filter with an epifluorescence microscope.Video 1. Visualization of the protocol as a video recording of the procedures
Data analysis
Representative results: The protocol described above provides an efficient enzymatic way to obtain B. graminis haustoria from the host plant, barley. All materials obtained using this protocol, including epiphytic mycelia-contained acetate stripes, haustoria-present epidermal peels and purified haustoria, can be stored at -80 °C for long-term storage till needed for further analysis. One pot of heavily infected barley (roughly 80 leaves) yields approximately 1.5 g of epiphytic material (N.B. this includes the cellulose acetate film, which constitutes the major part of this fraction, by weight), 0.3 g of epidermis and 3-4 x 105 haustoria (the actual number of haustoria obtained will depend strongly on the extent to which the leaves were infected). The quality of isolated haustoria can be observed by epifluorescence microscopy after staining by WGA-Alexa Fluor 488. Most of the isolated haustoria are highly branched and intact (Figure 1A). Sometimes the separated haustorium is still surrounded by the plant-derived extrahaustorial membrane, as shown in Figure 1B (arrow). Plant fragments which are small enough can also pass through the 70 μm nylon mesh sieve, so limited amount of impurities are present in pelleted haustoria complex, for example chloroplast (Figure 2, arrow).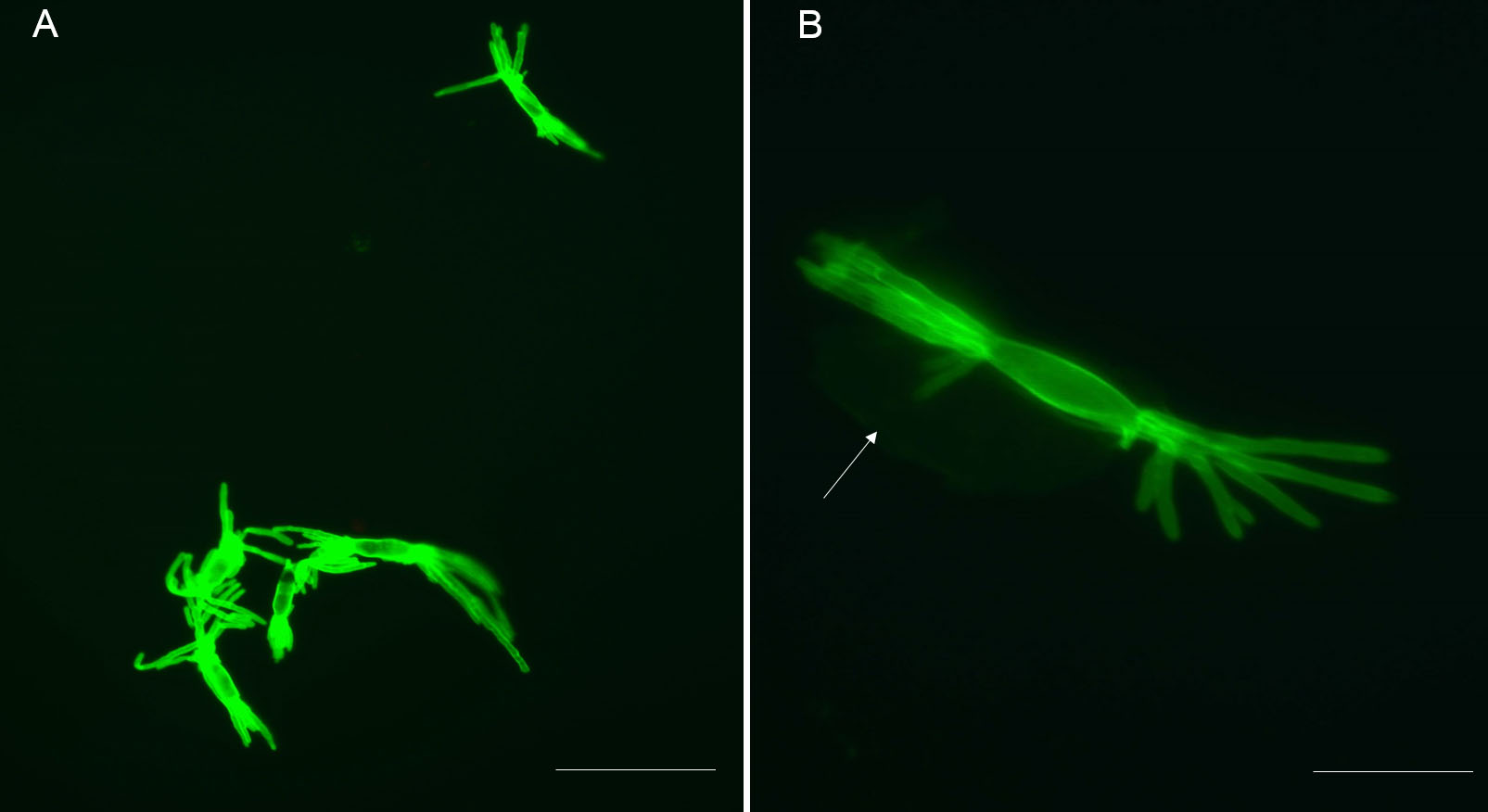
Figure 1. Isolated haustoria visualized after staining with WGA-Alexa Fluor 488. A. Stained haustoria (fluorescent green) visualized by epifluorescence microscopy and observed through a “GFP” filter. Multiple intact and mature haustoria. Scale bar = 40 μm. B. This shows both the multidigitated haustorium and a perihaustorial membrane surrounding the haustorium (arrow). Scale bars = 10 μm.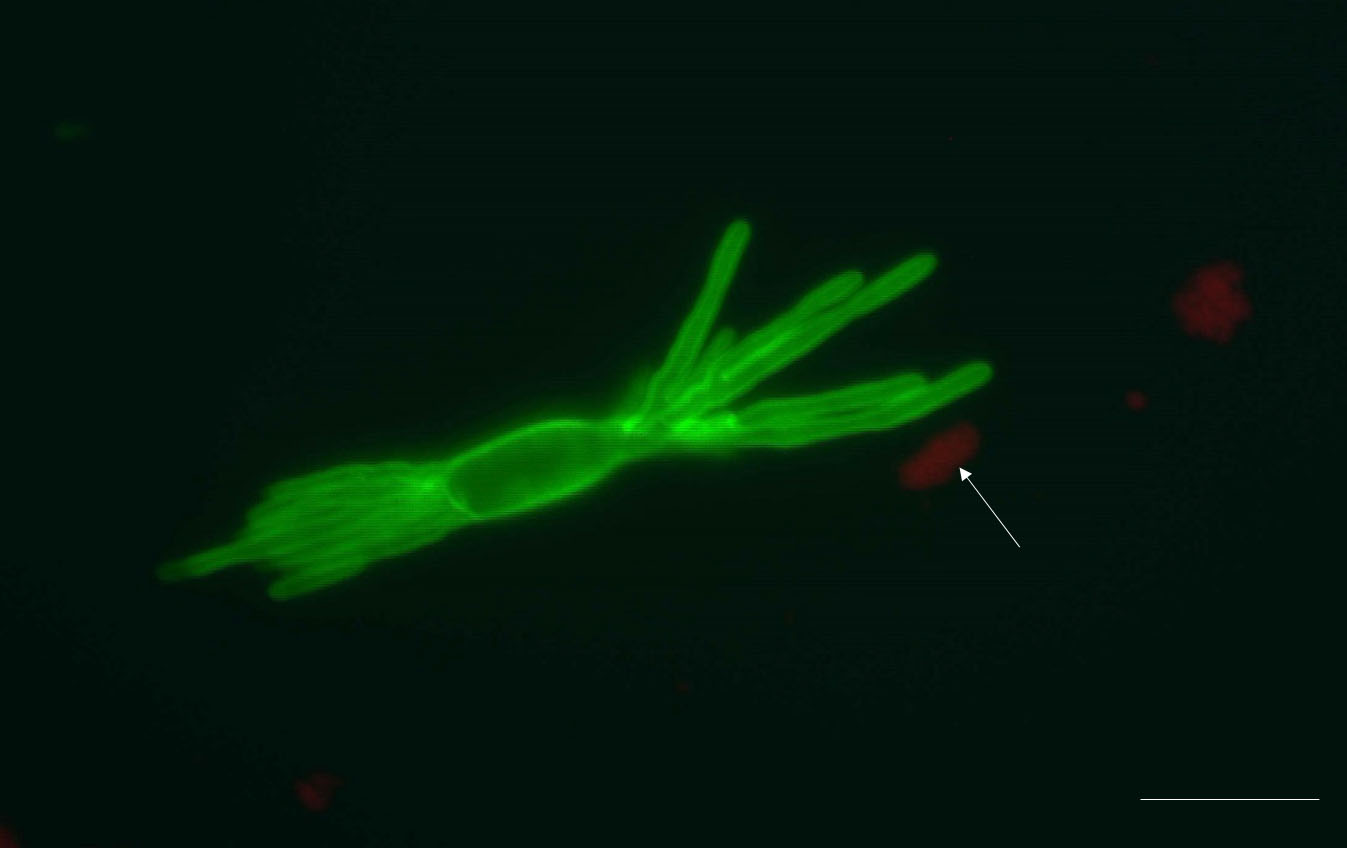
Figure 2. An isolated haustorium with some impurities. The pellet formed after filtration and centrifugation not only consists of haustoria, but also some plant debris, for example chloroplasts (autofluorescent red, arrow). These impurities could be chloroplasts and cell wall fragments. Scale bar = 10 μm.
We tested several options for overall digestion time in cellulase: 1 h, 2 h, 3 h and overnight. Overnight incubation yielded the highest number of haustoria: approximately 5 x 105 (this was considered the maximum amount of haustoria we can achieve), but also more fragments of plant material which reduced the purity of isolated haustoria. One-hour incubation only released less than 30% of the maximum obtained after overnight digestion. Digestions of 2 h and 3 h incubation yielded 3-4 x 105 haustoria, which accounts for > 80% of the maximum.
Discussion: Powdery mildew fungi are challenging to study because they have an absolute requirement for a host plant to grow and develop. This is especially true of the haustoria, the feeding structures which develop inside the host plant epidermal cells (Catanzariti et al., 2006; Micali et al., 2011). Several affinity isolation methods of powdery mildew haustoria have been previously published, including one modified method on B. graminis (Mackie et al., 1991; Micali et al., 2008). However, the method demonstrated for B. graminis, which combined gradient centrifugation and multiple filtrations, produce a pellet with 80% haustoria; the rest were reported to be cell wall fragments and chloroplasts (Godfrey et al., 2009). Isolating protoplast from different plants in previous studies is mostly achieved by a combination of enzymes, including cellulose (Sun et al., 2013; Wu et al., 2017). To obtain relatively pure and intact haustoria from barley powdery mildew, we developed a rapid and effective method which relies on the dissection of infected epidermis from the rest of the leaf followed by degradation of the plant cell walls and lysis of the epidermal cells using cellulase.
Maximizing the number of haustoria present in infected leaves is important when developing the protocol. Barley cv. Golden Promise was chosen to maintain the fungi due to its high susceptibility to powdery mildew. An appropriate number of seeds were sown to ensure there are enough plants while the density of the seedling is not too high to interfere with a saturating inoculation of the primary leaves. In these studies, we used approximately 3 g of seeds for a 10 cm-diameter pot. In order to ensure a high infection rate, conidia that are older than one day need to be removed from the plants used as a source of the inoculum. In this way, we ensure that only fresh conidia (< 1 day old) are used, leading to high percentage of successful germination, penetration and finally haustoria formation. The effectiveness of epidermis dissection is critical for the success of haustoria isolation. The young leaves are easier to dissect: there is a trade-off between more advanced stages of infection (yielding more haustoria) and younger leaves (yielding more intact epidermis). In practice, we have found that using plants 7 days post inoculation (dpi) as an ideal compromise. In addition, the epidermis of the abaxial side of primary leaf is the easiest to dissect compared to either the adaxial epidermis, or to the epidermis of other leaves. We therefore routinely dissect only the abaxial epidermis of the primary leaves.
To optimize the condition of haustoria isolation, several different incubation times and enzyme concentrations have been tested. Increasing concentration of cellulase (higher than 2.5%) and a combination of various enzymes (cellulase and macerozyme) have been proven effective in degrading cell walls, however, they were found to be costly and led to low viability of isolated structures (Wu et al., 2017). Accordingly, we used 2% of cellulase to break down the cell walls for releasing haustoria. The time of incubation in cellulase was critical to obtain high yield of isolated haustoria and low contamination from plant derived structures. After comparing different incubation times, we suggest that 2 h is a suitable compromise, for most purposes.
This protocol utilized WGA-AF 488 to stain isolated haustoria rather than the traditional method of using Coomassie blue. The lectin moiety binds to fungal chitin and rendered the fungal structures visible as green fluorescent structures due to the Alexa 488 fluorophore clearly distinguishable from plant derived material. If required, it would be possible to visualize the contaminating plant cell wall residues by staining with Calcofluor White.
We believe this protocol may be applied to other powdery mildews with suitable modifications.
Recipes
- 1x PBS buffer
8 g NaCl
0.2 g KCl
1.44 g Na2HPO4
0.24 g KH2PO4
Dissolved in 800 ml distilled water
Adjust the pH to 7.4 with HCl
Add distilled water to a total volume of 1 L
Acknowledgments
LL was funded by the China Scholarship Council (CSC) studentship, and BC was funded by The Gregory Trust (Imperial College London).
Competing interests
The authors declare that there are no competing interests in the work described here.
References
- Bindschedler, L. V., Burgis, T. A., Mills, D. J., Ho, J. T., Cramer, R. and Spanu, P. D. (2009). In planta proteomics and proteogenomics of the biotrophic barley fungal pathogen Blumeria graminis f. sp. hordei. Mol Cell Proteomics 8(10): 2368-2381.
- Bindschedler, L. V., McGuffin, L. J., Burgis, T. A., Spanu, P. D. and Cramer, R. (2011). Proteogenomics and in silico structural and functional annotation of the barley powdery mildew Blumeria graminis f. sp. hordei. Methods 54(4): 432-441.
- Bindschedler, L. V., Panstruga, R. and Spanu, P. D. (2016). Mildew-omics: how global analyses aid the understanding of life and evolution of powdery mildews. Front Plant Sci 7: 123.
- Both, M., Csukai, M., Stumpf, M. P. and Spanu, P. D. (2005a). Gene expression profiles of Blumeria graminis indicate dynamic changes to primary metabolism during development of an obligate biotrophic pathogen. Plant Cell 17(7): 2107-2122.
- Both, M., Eckert, S. E., Csukai, M., Müller, E., Dimopoulos, G. and Spanu, P. D. (2005b). Transcript profiles of Blumeria graminis development during infection reveal a cluster of genes that are potential virulence determinants. Mol Plant Microbe Interact 18(2): 125-133.
- Catanzariti, A. M., Dodds, P. N., Lawrence, G. J., Ayliffe, M. A. and Ellis, J. G. (2006). Haustorially expressed secreted proteins from flax rust are highly enriched for avirulence elicitors. Plant Cell 18(1): 243-256.
- Garnica, D. P., Nemri, A., Upadhyaya, N. M., Rathjen, J. P. and Dodds, P. N. (2014). The ins and outs of rust haustoria. PLoS Pathog 10(9): e1004329.
- Godfrey, D., Zhang, Z., Saalbach, G. and Thordal-Christensen, H. (2009). A proteomics study of barley powdery mildew haustoria. Proteomics 9(12): 3222-3232.
- Mackie, A. J., Roberts, A. M., Callow, J. A. and Green, J. R. (1991). Molecular differentiation in pea powdery-mildew haustoria: Identification of a 62-kDa N-linked glycoprotein unique to the haustorial plasma membrane. Planta 183(3): 399-408.
- Micali, C., Gollner, K., Humphry, M., Consonni, C. and Panstruga, R. (2008). The powdery mildew disease of Arabidopsis: A paradigm for the interaction between plants and biotrophic fungi. Arabidopsis Book 6: e0115.
- Micali, C. O., Neumann, U., Grunewald, D., Panstruga, R. and O'Connell, R. (2011). Biogenesis of a specialized plant-fungal interface during host cell internalization of Golovinomyces orontii haustoria. Cell Microbiol 13(2): 210-226.
- Pedersen, C., Ver Loren van Themaat, E., McGuffin, L. J., Abbott, J. C., Burgis, T. A., Barton, G., Bindschedler, L. V., Lu, X., Maekawa, T., Wessling, R., Cramer, R., Thordal-Christensen, H., Panstruga, R. and Spanu, P. D. (2012). Structure and evolution of barley powdery mildew effector candidates. BMC Genomics 13: 694.
- Pennington, H. G., Li, L. and Spanu, P. D. (2016). Identification and selection of normalization controls for quantitative transcript analysis in Blumeria graminis. Mol Plant Pathol 17(4): 625-633.
- Pliego, C., Nowara, D., Bonciani, G., Gheorghe, D. M., Xu, R., Surana, P., Whigham, E., Nettleton, D., Bogdanove, A. J., Wise, R. P., Schweizer, P., Bindschedler, L. V. and Spanu, P. D. (2013). Host-induced gene silencing in barley powdery mildew reveals a class of ribonuclease-like effectors. Mol Plant Microbe Interact 26(6): 633-642.
- Spanu, P. D., Abbott, J. C., Amselem, J., Burgis, T. A., Soanes, D. M., Stuber, K., Ver Loren van Themaat, E., Brown, J. K., Butcher, S. A., Gurr, S. J., Lebrun, M. H., Ridout, C. J., Schulze-Lefert, P., Talbot, N. J., Ahmadinejad, N., Ametz, C., Barton, G. R., Benjdia, M., Bidzinski, P., Bindschedler, L. V., Both, M., Brewer, M. T., Cadle-Davidson, L., Cadle-Davidson, M. M., Collemare, J., Cramer, R., Frenkel, O., Godfrey, D., Harriman, J., Hoede, C., King, B. C., Klages, S., Kleemann, J., Knoll, D., Koti, P. S., Kreplak, J., Lopez-Ruiz, F. J., Lu, X., Maekawa, T., Mahanil, S., Micali, C., Milgroom, M. G., Montana, G., Noir, S., O'Connell, R. J., Oberhaensli, S., Parlange, F., Pedersen, C., Quesneville, H., Reinhardt, R., Rott, M., Sacristan, S., Schmidt, S. M., Schon, M., Skamnioti, P., Sommer, H., Stephens, A., Takahara, H., Thordal-Christensen, H., Vigouroux, M., Wessling, R., Wicker, T. and Panstruga, R. (2010). Genome expansion and gene loss in powdery mildew fungi reveal tradeoffs in extreme parasitism. Science 330(6010): 1543-1546.
- Sun, H., Lang, Z., Zhu, L. and Huang, D. (2013). Optimized condition for protoplast isolation from maize, wheat and rice leaves. Sheng Wu Gong Cheng Xue Bao 29(2): 224-234.
- Testut, J. F., Callow, J. A. and Green, J. R. (1999). Evidence that PSI-D, a chloroplast photosystem I protein, is in haustoria of the powdery mildew fungus Erysiphe pisi. Physiological and Molecular Plant Pathology 55(6): 349-358.
- Wang, W., Wen, Y., Berkey, R. and Xiao, S. (2009). Specific targeting of the Arabidopsis resistance protein RPW8.2 to the interfacial membrane encasing the fungal haustorium renders broad-spectrum resistance to powdery mildew. Plant Cell 21(9): 2898-2913.
- Wu, J. Z., Liu, Q., Geng, X. S., Li, K. M., Luo, L. J. and Liu, J. P. (2017). Highly efficient mesophyll protoplast isolation and PEG-mediated transient gene expression for rapid and large-scale gene characterization in cassava (Manihot esculenta Crantz). BMC Biotechnol 17(1): 29.
Article Information
Copyright
© 2019 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Li, L., Collier, B. and Spanu, P. D. (2019). Isolation of Powdery Mildew Haustoria from Infected Barley. Bio-protocol 9(14): e3299. DOI: 10.21769/BioProtoc.3299.
Category
Plant Science > Plant immunity > Host-microbe interactions
Cell Biology > Cell isolation and culture > Cell isolation
Microbiology > Microbe-host interactions > Fungus
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









