- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Analysis of Functional Virus-generated PAMP RNAs Using IFNα/β ELISA Assay
Published: Vol 9, Iss 12, Jun 20, 2019 DOI: 10.21769/BioProtoc.3282 Views: 5531
Reviewed by: Vamseedhar RayaproluOmar AkilAmar Parvate

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

General Maintenance and Reactivation of iSLK Cell Lines
Ariana C. Calderón-Zavala [...] Ekaterina E. Heldwein
Jun 5, 2025 1876 Views

Inducible HIV-1 Reservoir Reduction Assay (HIVRRA), a Fast and Sensitive Assay to Test Cytotoxicity and Potency of Cure Strategies to Reduce the Replication-Competent HIV-1 Reservoir in Ex Vivo PBMCs
Jade Jansen [...] Neeltje A. Kootstra
Jul 20, 2025 2457 Views

Assembly and Mutagenesis of Human Coronavirus OC43 Genomes in Yeast via Transformation-Associated Recombination
Brett A. Duguay and Craig McCormick
Aug 20, 2025 3033 Views
Abstract
Virus-generated PAMP RNAs are key factors that activate host immune response. The PAMP RNAs are therefore usually closely related with viral disease pathogenesis. Quantitative real time PCR is a conventional method to assess RNA. However, it cannot be used for detecting short dsRNAs generated by viral replicase. This protocol was established to analyze the PAMP RNAs produced by viruses which are able to induce host immune response. Classical viral PAMP RNAs and non-classical viral PAMP RNAs are analyzed separately. Briefly, to access total viral PAMP RNAs, total RNA was extracted from the virus infected cells and then transfected into Cop5 cells. Whereas, to assess non-classical viral PAMP RNAs, the constructs expressing viral replicase are transfected into Cop5 cells. The amount of IFNα/β produced by Cop5 cells, determined by ELISA, is correlated with the total and non-classical viral PAMP RNAs. Since this method is based on type I IFN response, it is therefore suitable for measuring the functional virus-generated PAMP RNAs and also for assessing the efficiency of these PAMP RNAs.
Keywords: VirusBackground
Under virus infection, the activation of host immune system usually starts with the recognition of viral pathogen-associated molecular patterns (PAMPs) by host pattern recognition receptors (PRRs) (Medzhitov and Janeway, 2002). Viral PAMPs, associated with virus invasion, include virus surface glycoproteins, viral DNA and RNAs (Mogensen and Paludan, 2005). In case of RNA virus infection, virus replication generated RNAs are essential PAMPs portion which is able to be recognized by the host intracellular PRRs (Alexopoulou et al., 2001). Two possible candidates for the role of such viral-generated PAMP RNAs have been recently reported. First, classical viral PAMP RNAs are dsRNA replication forms that are recognized by RIG-I and MDA5 and non-capped positive-strand RNAs (polyA+) generated by alphaviruses (Sokoloski et al., 2015) that may be recognized by RIG-I (Akhrymuk et al., 2015). Second, non-classical viral PAMP RNAs are short non-polyadenylated dsRNAs with 5’ triphosphate. They are IFN-inducing RNAs generated by viral replicase using cellular RNA templates (Nikonov et al., 2013). In Semliki Forest virus (SFV) infected cells these RNAs represent the majority of PAMP RNAs (Nikonov et al., 2013).
Given the essential role of viral PAMP RNAs to activate host immune system, a robust protocol to analyze different types of virus-generated PAMP RNAs is useful for virus-host interaction study. Quantitative real time PCR could be used to quantify classical viral dsRNAs by targeting negative strands. However, it is hard to differentiate between capped and non-capped positive strands. Furthermore, the approach is neither able to tell if the targeted PAMP RNA is functional nor able to access the PAMP efficiency and cannot detect the non-classical viral PAMP RNAs. To analyze the functional PAMP RNAs that are able to trigger host immune response, we established this protocol to analyze both classical and non-classical viral PAMP RNAs. To assess the total viral PAMP RNAs, total RNA from the infected cells were extracted for the assessment. Since all types of classical alphaviral PAMP RNAs have unpaired polyA tails and they are presumably the biggest proportion in the viral PAMP RNAs, they may overshadow other types of PAMP RNAs. Therefore, in a parallel experiment, we separated and examined the polyA- fraction from the total RNAs (Nikonov et al., 2013). To assess the non-classical viral PAMP RNAs, viral replicase constructs were used for analysis because the system did not have replication competent viral RNA templates and therefore no classical viral PAMP RNAs were produced. Cop5 cells were used as IFNα/β activation host for all PAMP RNAs analyses. The quantity of the IFNα/β produced by the transfected PAMP RNAs can be determined by ELISA assay.
This protocol is a reliable method to analyze alphavirus-generated PAMP RNAs during the virus replication and, with exception of separation of polyA- fraction, applied to viruses that lack unpaired polyA tails. It is not only to measure the level of the PAMP RNAs but also is able to assess the PAMP RNAs efficiency. Therefore, this protocol is suitable for the study involving the interaction between RNA virus invasion and the host immune response.
Materials and Reagents
- Materials
- 1.5 ml Eppendorf® DNA LoBind microcentrifuge tubes (Eppendorf, catalog number: 0030108051)
- 100 mm Corning® tissue-culture treated culture dishes (Corning, catalog number: 430167)
- 12-well CorningTM CostarTM flat bottom cell culture plates (Corning, catalog number: 3513)
- 15 ml FalconTM conical centrifuge tubes (Corning, catalog number: 352096)
- 6-well plate
- Cell lines
- ATCC® BHK-21[C13] cells (ATCC, catalog number: CCL-10TM)
- Cop5 cells (Tyndall et al., 1981)
- Viruses of interest
Note: Prepare the viruses of interest from infectious clones. It is recommended that the virus titer should be prepared between 105 and 107 plaque forming units (PFU)/ml. The viruses listed below are for a demonstration. These viruses are derived from infectious clones, released (passage 0), passaged in BHK-21 cells and stored at -80 °C.
- RRV-T48
- RRV-T48A534V
- Viral replicase constructs
Note: Prepare the constructs expressing the viral replicase (non-structural proteins) of the viruses of interest. The constructs listed below are for a demonstration. As shown in the construct map below (Figure 1), the regions encoding the replicase of RRV-T48, RRV-T48A534V and SFV4 (used as positive control [Nikonov et al., 2013]) were cloned into pMC-gtGTU2 vector, which contains cytomegalovirus early promoter, leader sequence from the thymidine kinase of Herpes Simplex virus (HSV) with an inserted synthetic intron and late polyadenylation signal from simian virus 40 (SV40).
- RRV-T48
- RRV-T48A534V
- SFV
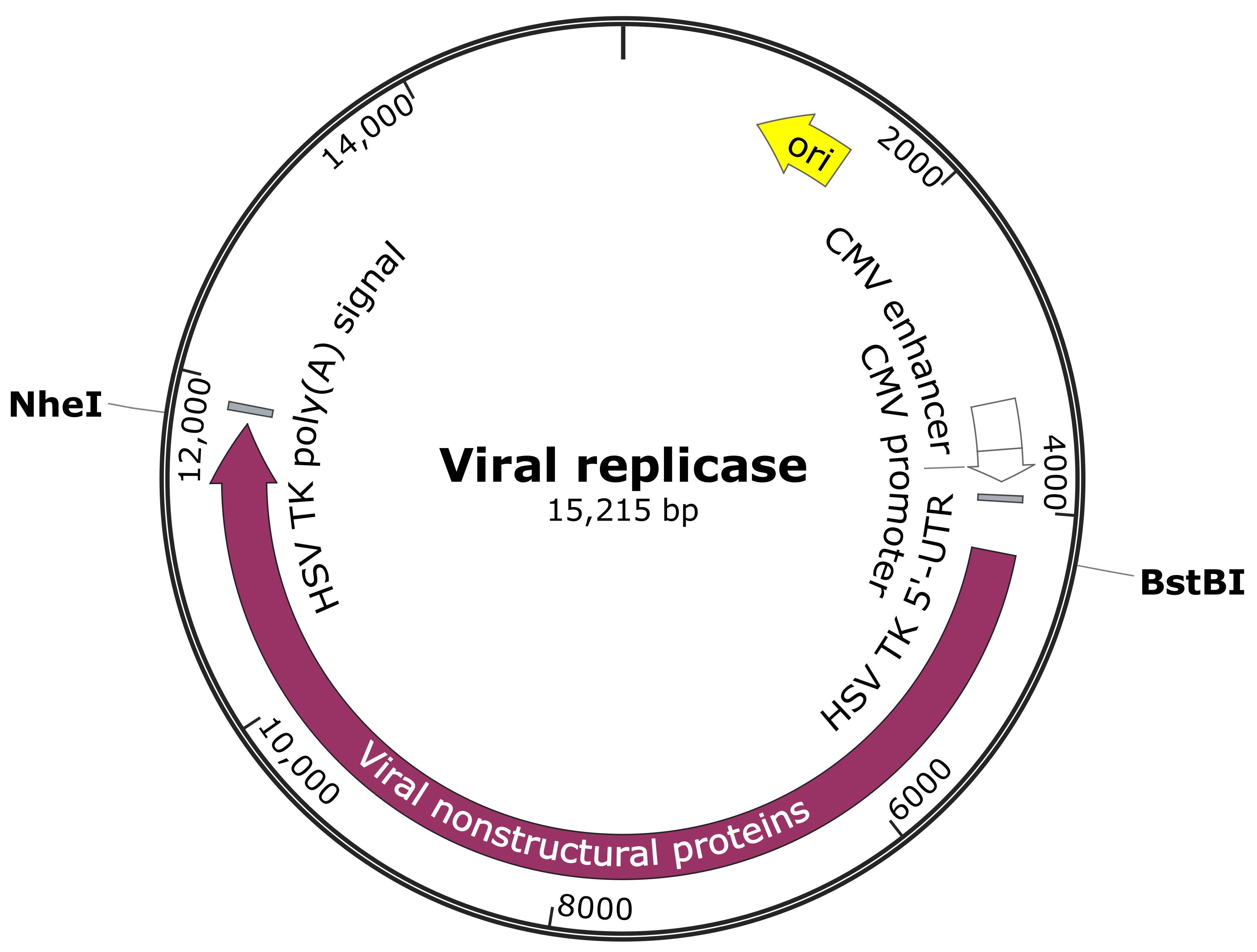
Figure 1. Viral replicase construct map
- Reagents
- Dulbecco's modified Eagle medium (DMEM) (Sigma-Aldrich, catalog number: D5030)
- Iscove’s Modified Dulbecco’s Medium (IMDM) (Sigma-Aldrich, catalog number: 51471C)
- L-Glutamine (Sigma-Aldrich, catalog number: G6392)
- Fetal bovine serum (FBS) (Sigma-Aldrich, catalog number: F6178)
- 100x Penicillin-streptomycin (Sigma-Aldrich, catalog number: P4333)
- Tryptose phosphate broth solution (TPB) (Sigma-Aldrich, catalog number: T8159)
- Phosphate buffered saline (PBS) (Sigma-Aldrich, catalog number: P5493)
- 1 M HEPES buffer (Sigma-Aldrich, catalog number: 83264)
- 0.25% Trypsin-EDTA solution (Sigma-Aldrich, catalog number: T4049)
- RNeasy Mini Kit (QIAGEN, catalog number: 74104)
- TRIzol® Reagent (Invitrogen, catalog number: 15596026)
- DNase I (Roche, catalog number: 04716728001)
- PolyATtract® mRNA Isolation Systems (Promega, catalog number: Z5210)
- LipofectamineTM 2000 reagent (Thermo Fisher Scientific, catalog number: 11668019)
- VeriKine Mouse Interferon Beta ELISA Kit (PBL Assay Science, catalog number: 42400-1)
- pMC-gtGTU2 vector (Nikonov et al., 2013) or any mammalian expression vector (such as pcDNA or pCMV series from Thermo Fisher Scientific)
Equipment
- -80 °C freezer
- 4 °C refrigerator
- Vortexer (Bio-Rad)
- Pipettes (Bio-Rad, catalog numbers: 166-0506, 166-0507 and 166-0508)
- CO2 incubator (Bio-Rad, catalog number: 4110)
- Biosafety level 2 (BSL-2) cabinet
- Autoclave (Tuttnauer, ELV-D Line)
- UVC5000 Ultraviolet Crosslinker (Hoefer, code number: UVC5000-230V)
- SunriseTM absorbance microplate reader (Tecan)
- Microfuge 16 Centrifuge (Beckman Coulter Life science, catalog number: A46473)
Software
- Microsoft Excel or Prism 5
Procedure
The procedure showing the analysis scheme for viral PAMP RNAs is summarized in Figure 2.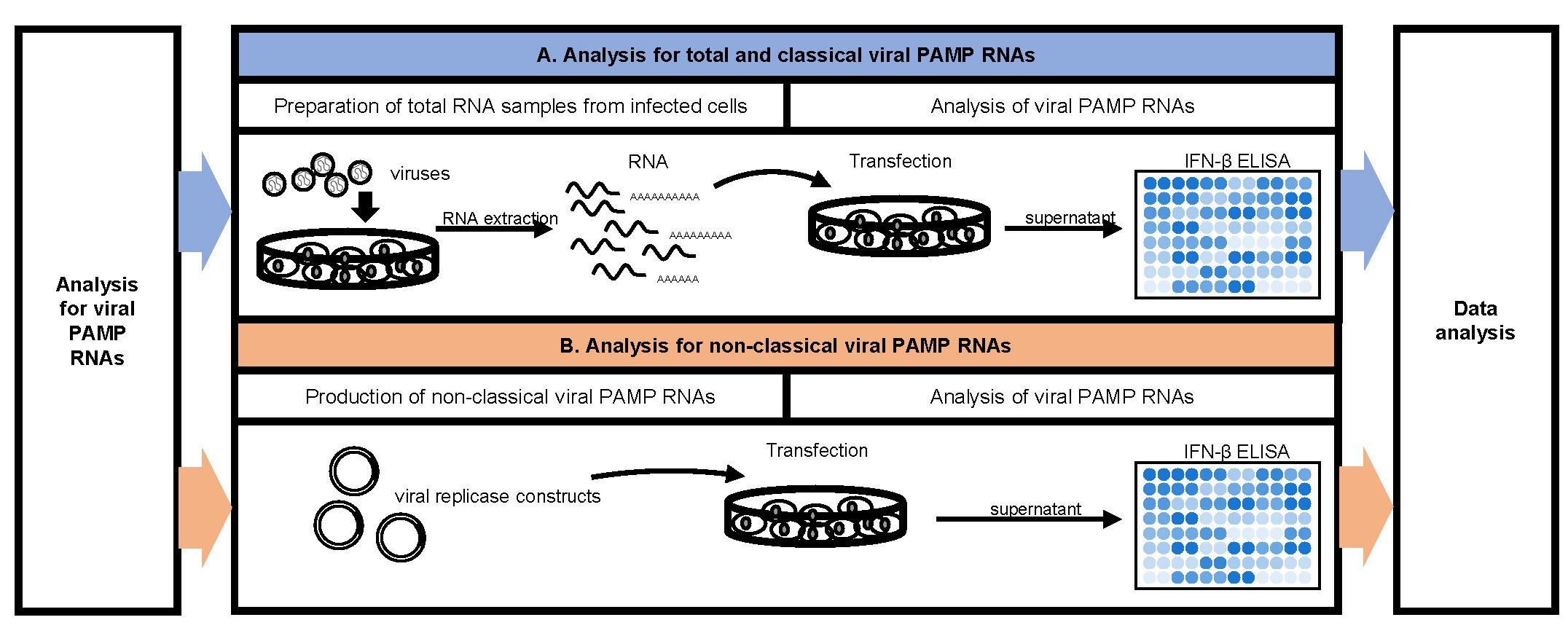
Figure 2. Flow chart for viral PAMP RNAs analysis
- Analysis for total and classical viral PAMP RNAs
- Preparation of total RNA samples from infected cells.
Day -1
- Seed 8 x 106 BHK-21 cells on to 100 mm cell culture dish and culture in DMEM with 10% FBS. Maintain the cells at 37 °C with 5% CO2 overnight.
- Discard the BHK-21 cell growth media and wash the cell monolayer once with PBS. Infect the cells with the viruses of interest. As a demonstration, dilute RRV-T48 and RRV-T48A534V separately in serum free DMEM for the infection. Infect the BHK-21 cell monolayer by the viruses at Multiplicity of Infection (MOI) 1.0 with a minimum coverage volume (approximately 400 μl for each well in a 6-well plate). Incubate the cells for 1 h at 37 °C with 5% CO2 and then maintain the cells in DMEM with 10% FBS for another 6 h.
- Extract the total RNA from the infected BHK-21 cells using TRIzol Reagent according to the manufacturer’s protocol (Invitrogen TRIzolTM Reagent USER GUIDE). Use 1 U DNase I per 1 μg total RNA to treat the extracted RNA for 60 min at 37 °C.
- Optional (viruses with non-paired polyA tail): Isolation of non-classical PAMPs. Aliquot 100 μg of the treated RNA into a clean Eppendorf® DNA LoBind microcentrifuge tube (RNase free). Remove the polyA(+) fraction from the total RNA using PolyATtract mRNA Isolation Systems Kit according to the manufacturer’s protocol (Promega).
- Purify the total, polyA(+) and polyA(-) RNA samples with RNeasy Mini Kit according to the manufacturer’s protocol (Qiagen) and elute the product within 20 μl RNase free water.
- UV inactivates the infectious viral RNAs, presented in the obtained samples, for 5 min in a UVC5000 Ultraviolet Crosslinker at 2000 μJ/cm².
- Optional: Store the RNA at -80 °C for longer period if needed.
- Seed 8 x 106 BHK-21 cells on to 100 mm cell culture dish and culture in DMEM with 10% FBS. Maintain the cells at 37 °C with 5% CO2 overnight.
- Analysis of viral PAMP RNAs
Day -1
- Seed 1 x 106 Cop5 cells into 12-well plate and cultured in IMDM with 10% FBS and 2 mM L-glutamine. Maintain the cells at 37 °C with 5% CO2 overnight.
- Transfect 1 μg of treated total, polyA(+) or polyA(-) RNAs respectively into confluent cultures of Cop5 cells in 12-well plates using Lipofectamine2000 reagent. Briefly, dilute 3 μl of the Lipofectamine2000 reagent in 100 μl serum free IMDM and incubate for 2-5 min at room temperature. Dilute 1 μg RNA samples in 100 μl serum free IMDM and incubate for 2-5 min at room temperature. Combine the 100 μl diluted Lipofectamine2000 reagent with the 100 μl diluted RNA samples and incubate the RNA-lipid mixture for 5 to 10 min at room temperate to form the RNA-lipid complex. Discard the BHK-21 cell growth media and wash the cell monolayer once with PBS. Add 1 ml serum free IMDM to the cells. Add the RNA-lipid complex drop-wise on top of the medium and gently rock the plate after adding to the mix. Incubate the transfected cells at 37 °C with 5% CO2 for 24 h.
- Collect the supernatant from the transfected Cop5 cells to a 1.5 ml tube. Centrifuge samples at 3,000 x g for 5 min at 4 °C to remove the cell debris. Collect the supernatant to a new tube.
- Determine the amount of IFN-β produced by the transfected Cop5 cells using VeriKine Mouse IFN Beta ELISA kit according to the manufacturer’s protocol together with standard curve.
- Preparation of total RNA samples from infected cells.
- Analysis for non-classical viral PAMP RNAs
Day -1- Seed 1 × 106 Cop5 cells into a 12-well plate and culture in IMDM with 10% FBS and 2 mM L-glutamine. Maintain the cells at 37 °C with 5% CO2 overnight.
- Transfect 1 μg of the plasmids encoding for viral replicase of interest into the confluent cultures of Cop5 cells in 12-well plates using Lipofectamine2000 reagent. Use plasmid that expresses inactive replicase (active-site mutant) as a negative control. As a demonstration, transfect RRV-T48, RRV-T48A534V and SFV4 replicase constructs into the Cop5 cells for the following analysis.
- Incubate the transfected cells at 37 °C with 5% CO2 for 48 h.
- Collect the supernatant from the transfected Cop5 cells to a 1.5 ml tube. Centrifuge samples at 3,000 x g for 5 min at 4 °C to remove the cell debris. Collect the supernatant to a new tube.
- Determine the amount of IFN-β produced by the transfected Cop5 cells using VeriKine Mouse IFN Beta ELISA kit according to the manufacturer’s protocol (pbl Assay science) together with standard curve. For each independent experiment, normalized the IFN-β levels measured in supernatants from cells transfected with RRV replicase expression plasmids to the amount of IFN-β produced by cells transfected by plasmid expressing SFV replicase.
Data analysis
Collect the data of the amount of IFN-β in Microsoft Excel or Prism 5. Analyze the data using Student’s two-tailed unpaired t-test. As a demonstration, the standard curve for IFN-β ELISA assay was calculated in Table 1. The levels of IFN-β produced by Cop5 cells under the transfection of total viral PAMP RNAs (Figure 3), non-classic viral PAMP RNAs (Figure 4) and the replicase constructs (Figure 5) from RRV-T48 or RRV-T48A534V were plotted in bar graphs. The corresponding plotting raw data were demonstrated in Tables S1, S2 and S3.
Table 1. IFN-β ELISA assay standard curve. Briefly, seven concentrations of the mouse IFN-β standard were prepared according to the following table. One-hundred microliters per well of interferon standard was added to each well and incubate for 1 h at room temperature. The liquid was removed and washed with wash solution for three times. One-hundred microliters of diluted antibody solution was added to each well and incubated for 1 h at room temperature. The contents in the wells were removed and washed with wash solution for three times. One-hundred microliters of diluted HRP Solution was added to the wells and incubated for 1 h at room temperature. The contents in the wells were removed and washed with wash solution for three times. One-hundred microliters of the TMB substrate solution was added to the wells and incubated for 15 min at room temperature in the dark. 100 μl of stop solution was added to each well to stop the reaction. The absorbance at 450 nm was determined within 5 min.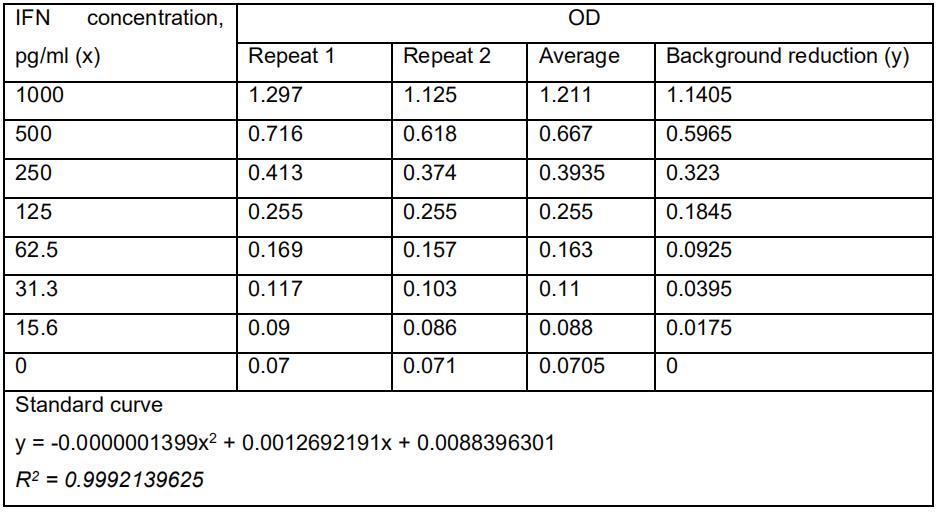
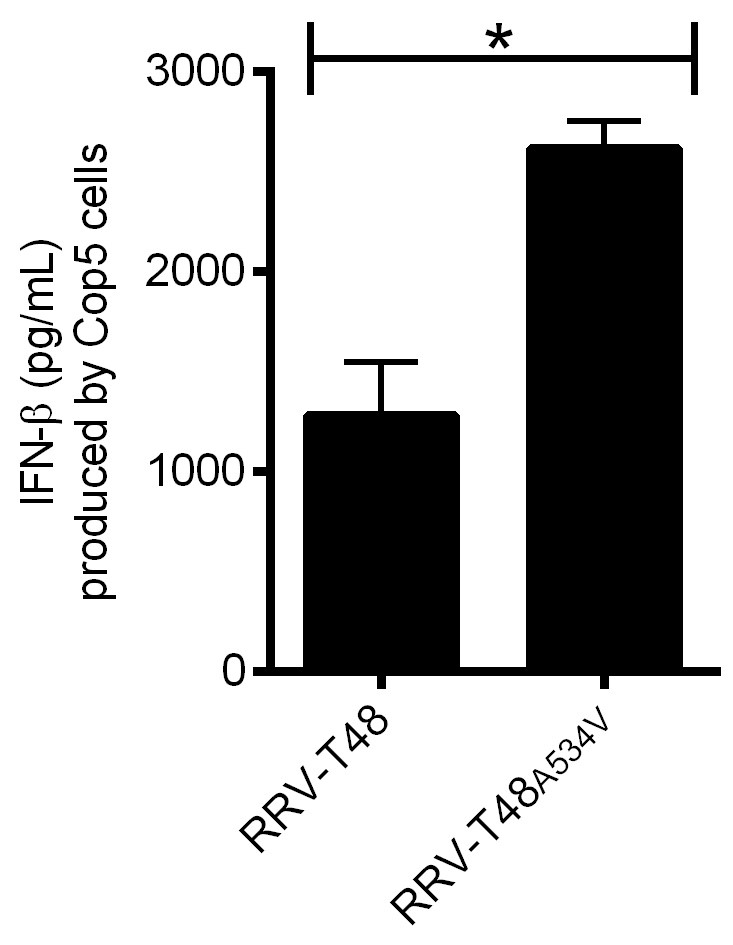
Figure 3. IFN-β produced by Cop5 cells by the transfection of total viral PAMP RNAs from RRV-T48 or RRV-T48A534V. BHK-21 cells were infected with RRV-T48 and RRV-T48A534V separately at MOI 1.0. Cells were lysed and total RNA was isolated at 6 h post infection. One microgram of each RNA sample was used for transfection of Cop5 cells. The amount of IFN-β in the cell supernatant at 24 h post transfection was determined. IFN-β levels were expressed as the means ±SEM from three experiments (*, P < 0.05 using Student’s two-tailed unpaired t-test).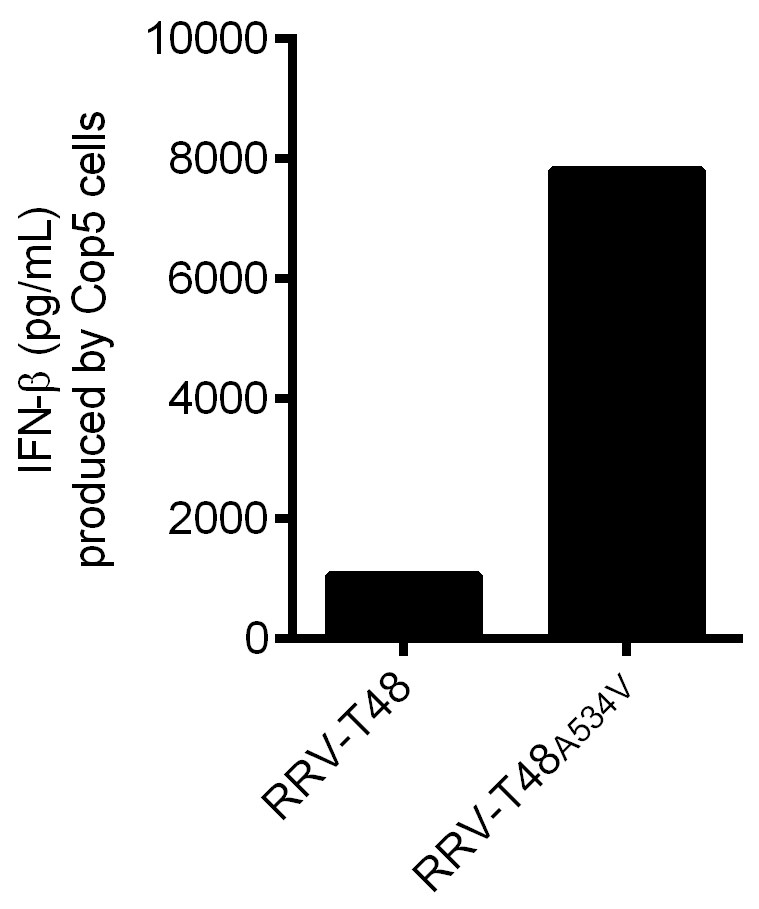
Figure 4. IFN-β produced by Cop5 cells by the transfection of non-classic viral PAMP RNAs from RRV-T48 or RRV-T48A534V. BHK-21 cells were infected with RRV-T48 and RRV-T48A534V separately at MOI 1.0. Cells were lysed and total RNA was isolated at 6 h post infection. Poly(A)- fraction was obtained from isolated RNAs by removal of poly(A)+ RNAs, including viral dsRNA replication forms. 1 μg of each RNA sample was used for transfection of Cop5 cells. The amount of IFN-β in the cell supernatant at 24 h post transfection was determined. 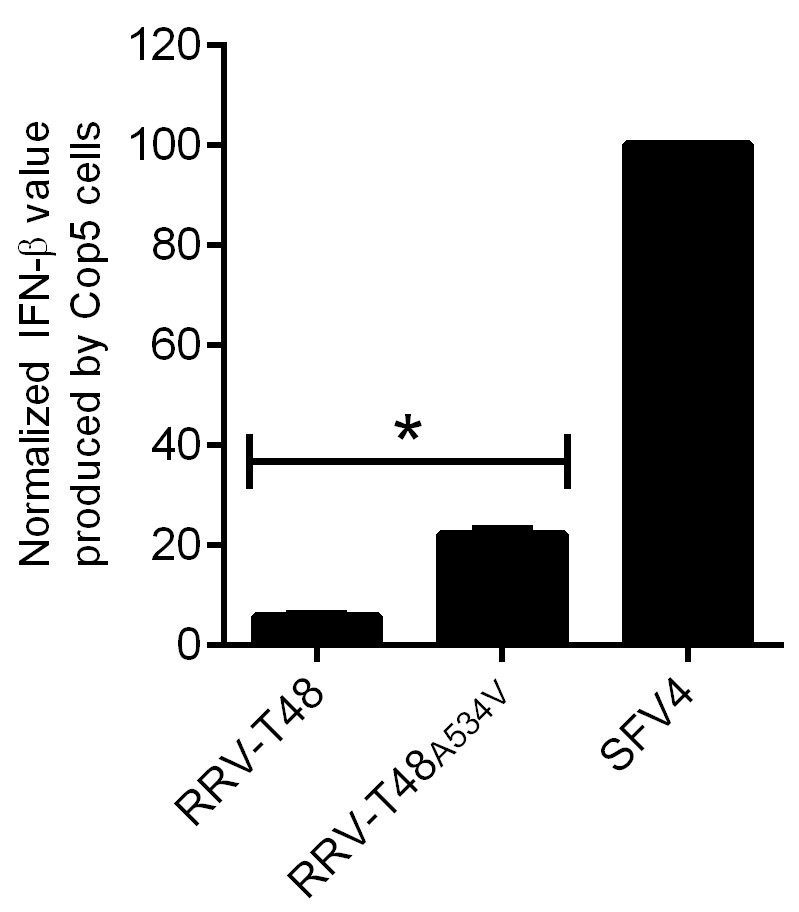
Figure 5. IFN-β produced by Cop5 cells by the transfection of replicase of RRV-T48 or RRV-T48A534V. Cop5 cells were transfected with plasmids designed to express RRV-T48, RRV-T48A534V or SFV4. The amount of IFN-β in supernatant was determined at 48 h post transfection. Values obtained for cells transfected with RRV-T48 or RRV-T48A534V replicase expression plasmids were normalized to values obtained for cells transfected with plasmid expressing SFV4 replicase (taken as 100). Normalized values were shown as the means ±SEM from three experiments (*, P < 0.05 using Student’s two-tailed unpaired t-test).
Acknowledgments
This protocol was originally published in the paper by Nikonov et al., 2013. This study was supported by grants from the Australian National Health and Medical Research Council to SM (APP1031024) and Estonian Research Council Grant to AM (IUT20-27). Suresh Mahalingam is the recipient of the NHMRC Senior Research Fellowship (ID: APP11544347).
Competing interests
All authors declare no competing interests.
References
- Akhrymuk, I., Frolov, I. and Frolova, E. I. (2015). Both RIG-I and MDA5 detect alphavirus replication in concentration-dependent mode. Virology 487: 230-241.
- Alexopoulou, L., Holt, A. C., Medzhitov, R. and Flavell, R. A. (2001). Recognition of double-stranded RNA and activation of NF-κB by Toll-like receptor 3. Nature 413(6857): 732-738.
- Medzhitov, R. and Janeway, C. A., Jr. (2002). Decoding the patterns of self and nonself by the innate immune system. Science 296: 298-300.
- Mogensen, T. H. and Paludan, S. R. (2005). Reading the viral signature by Toll-like receptors and other pattern recognition receptors. J Mol Med (Berl) 83(3): 180-192.
- Nikonov, A., Molder, T., Sikut, R., Kiiver, K., Mannik, A., Toots, U., Lulla, A., Lulla, V., Utt, A., Merits, A. and Ustav, M. (2013). RIG-I and MDA-5 detection of viral RNA-dependent RNA polymerase activity restricts positive-strand RNA virus replication. PLoS Pathog 9(9): e1003610.
- Tyndall, C., La Mantia, G., Thacker, C.M., Favaloro, J., Kamen, R., (1981). A region of the polyoma virus genome between the replication origin and late protein coding sequences is required in cis for both early gene expression and viral DNA replication. Nucleic Acids Research 9, 6231-6250.
- Sokoloski, K. J., Haist, K. C., Morrison, T. E., Mukhopadhyay, S. and Hardy, R. W. (2015). Noncapped alphavirus genomic RNAs and their role during infection. J Virol 89(11): 6080-6092.
Article Information
Copyright
© 2019 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Mutso, M., Liu, X., Merits, A. and Mahalingam, S. (2019). Analysis of Functional Virus-generated PAMP RNAs Using IFNα/β ELISA Assay. Bio-protocol 9(12): e3282. DOI: 10.21769/BioProtoc.3282.
Category
Microbiology > Microbe-host interactions > Virus
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link