- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Visualization of Macropinocytosis in Prostate Fibroblasts
Published: Vol 9, Iss 10, May 20, 2019 DOI: 10.21769/BioProtoc.3235 Views: 6889
Reviewed by: Mauro Sbroggio'Anonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Atomic Force Microscopy to Characterize Ginger Lipid-Derived Nanoparticles (GLDNP)
Dingpei Long [...] Didier Merlin
Apr 5, 2021 5974 Views

Modeling the Nonlinear Dynamics of Intracellular Signaling Networks
Oleksii S. Rukhlenko and Boris N. Kholodenko
Jul 20, 2021 4079 Views

Developing Clinically Relevant Acquired Chemoresistance Models in Epithelial Ovarian Cancer Cell Lines
Priti S. Shenoy [...] Pritha Ray
Feb 5, 2022 4213 Views
Abstract
Macropinocytosis has emerged as an important mechanism for non-selective route to internalize extracellular fluids and dissolved molecules in eukaryotic cell. As fundamental cellular behavior, macropinocytosis plays specific and distinct roles in many physiological and pathological processes, such as nutrients uptake, antigen presentation, pathogen capture, and tumorigenesis. It supports tumorigenesis by providing metabolic needs to dividing cells in Ras driven cancer. In recent years, macropinocytosis has gained considerable interest in physiology and various diseases, including cancer, neurodegenerative diseases and atherosclerosis, which in turn has led to the discovery of new endocytic recycling systems. Approaches to assess macropinocytosis will provide insight into its underlying regulatory molecular mechanisms and enable the physiological control of macropinocytosis for controlled drug delivery and targeted cancer therapy. Macropinocytosis is an important phenomenon in Ras-expressing cancer cells and, recently, we have revealed a functional role for macropinocytosis in cancer associated fibroblasts (CAFs) fueling cancer cell growth. Here, we describe a protocol for detection of macropinocytosis in prostatic fibroblasts in vitro by utilizing fluorescently-labeled, lysine-fixable, 70 kDa high molecular weight dextran. Macropinosomes are visualized as fluorescent intracellular puncta either by confocal or fluorescent microscopy. To follow, subsequent intracellular events and their underlying mechanisms after macropinosomes formation, we perform co-localization of quenched BSA (DQTM-BSA) along with dextran labeling in cancer associated fibroblasts. Our protocol provides a consistent way to understand macropinocytosis in wild type or genetic manipulated prostatic fibroblast.
Keywords: MacropinosomesBackground
Macropinocytosis or ‘cell drinking’ is a type of endocytosis that involves the nonspecific uptake of extracellular fluid into large intracellular vesicles known as macropinosomes (Swanson and Watts, 1995). The macropinosomes are heterogeneous in size and shape, with their diameters ranging between 0.2 and 5 micrometers (Lim and Gleeson, 2011). The Src and Ras are prominent oncogenes which stimulate macropinocytosis in various type of cancer (Bar-Sagi and Feramisco, 1986). Macropinocytosis has been prominently described in pancreatic cancer cells, recognized for having active Ras signaling (Commisso et al., 2013; Davidson et al., 2017). We determined that epigenetic activation of Ras signaling mediates macropinocytosis in prostatic cancer associated fibroblasts resulting in albumin uptake, lysosomal degradation, and release of constituent amino acids (Mishra et al., 2018). These amino acids, predominantly glutamine, were found to support the metabolism and differentiation of the prostate cancer epithelial cells. Therefore, macropinocytosis is an important nutrient processing pathway to support the energetic needs of cancer associated fibroblasts (CAFs) and may represent a predictive biomarker for cancer therapy. The quantification of macropinocytosis can be achieved through confocal laser scanning microscopy.
Materials and Reagents
- Pipette tips (Fisher Scientific, catalog number: 02707404)
- Sterile 10 ml serological pipette (Santa Cruz Biotechnology Inc., catalog number: sc-200281)
- Sterile 15 ml conical tubes (BD Falcon, catalog number: 669993)
- 12-well cell culture plate (Fisher Scientific, catalog number: 64976)
- Cover glass (Fisher Scientific, catalog number: 22050221)
- Aluminum foil (Fisher healthcare, catalog number: 01213101)
- Kimwipe (lint-free paper towel; Fisher healthcare, catalog number: S47299)
- Human prostatic fibroblasts and RasV12 (Mishra et al., 2018)
Note: These cell types have active Ras activity compared to their normal counterpart. - Fetal Bovine Serum (Atlanta Biologicals, catalog number: S11550)
- DMEM/F12 medium (Fisher Scientific, catalog number: SH300404)
- Tetramethylrhodamine-conjugated high molecular weight dextran (TMR-dextran, 75 kDa), lysine fixable (Invitrogen Molecular Probes, catalog number: D1818)
- DQ-BSA (Thermo Fisher Scientific, InvitrogenTM, catalog number: D12050)
- LysoTracker® Green DND-26 (Life Technologies, catalog number: L7526)
- 5-(N-ethyl-N-isopropyl) amiloride (EIPA) (Invitrogen Molecular Probes, catalog number: e-3111)
- Vectashield Antifade Mounting Medium with DAPI (Vector Laboratories, catalog number: H-1200)
- Formaldehyde solution, ACS reagent grade, 37% (vol/vol) (Sigma, catalog number: 252549)
- NuSerum (Fisher Scientific, catalog number: 355500)
- Insulin, human recombinant, zinc solution (Invitrogen, catalog number: 12585-014)
- Sodium chloride (NaCl) (Fisher Biosciences, catalog number: BP358-1)
- Dimethyl sulfoxide (DMSO) (Fisher Biosciences, catalog number: D128-1)
- Sodium dihydrogen phosphate (NaH2PO4) (Fisher Biosciences, catalog number: ICN19550080)
- Sodium Phosphate, Dibasic, Anhydrous, Na2HPO4 (Fisher Biosciences, catalog number: AC424375000)
- 10-8 M testosterone (Nacalai Tesque, catalog number: 32811-61)
- Poly-L-Lysine (Sigma, catalog number: P4707)
- EIPA (5-(N-ethyl-N-isopropyl) amiloride) (Sigma, catalog number: A3085)
- Sterile distilled water
- Stromal complete medium (see Recipes)
- PBS 1x, pH 7.4 (see Recipes)
- Dextran stock solution (see Recipes)
- EIPA stock solution (see Recipes)
- LysoTracker green DND-26 (see Recipes)
- DQ-Green BSA stock solution (see Recipes)
- Dextran cell culture incubation medium (see Recipes)
- Fixation buffer (see Recipes)
Equipment
- Pipette-aid
- Tissue culture hood
- Centrifuge with adaptors for 15 ml conical tubes
- Humidified cell culture incubator set to 37 °C and 5% CO2
- Confocal Laser Scanning Microscope (Leica, Germany)
- Fine-point forceps
- 4 °C refrigerator
- -20 °C refrigerator
Procedure
- Growing fibroblasts on coverslips
- Place the sterile cover slips in a 12-well plate; rinse the cover slips with Phosphate Buffered Saline (PBS), followed by a quick rinse with culture media. Plate the cells on the cover slips at a density of ~3,000/well in 500 µl of media.
- The labeling of macropinosomes involves cell growth under serum-starved conditions. Therefore, prior optimization is needed to find out suitable time point for getting 50%-75% confluency of cells. The confluency of the cell is very critical for proper labeling of macropinosomes and spreading on coverslip.
- Macropinosomes labeling
- Cells are cultured in a 5% CO2 incubator at 37 °C and all subsequent treatments are performed at 37 °C using pre-warmed media.
- Cells grown on cover slips in 12-well plates were serum starved for 18 h, followed by incubation with 1 mg/ml TMR-dextran for 30 min in serum free DMEM/F12 medium in CO2 incubator.
Note: Dextran is available in different colors and molecular weights and it can be selected as per requirement and desired goals of the experiment. - Remove culture medium as much as possible and wash the cells with 500 µl of ice cold PBS for 5 times. Fix the cells with 150 µl of 3.7% formaldehyde in PBS. Incubate the cells at room temperature for 20 min in the dark by covering it with aluminum foil.
Note: Proper rinsing of the samples is very crucial to avoid fluorescent background signal. - Remove the coverslip from the 12-well plates using forceps. Hold the coverslip vertical, with the bottom rim touching a paper towel to absorb remaining PBS and dry the coverslip completely.
- For nuclear staining and mounting, apply one drop of DAPI mounting medium to each glass slide. Take the cover slip and set it at an angle to the slide so that one edge of it touches DAPI mounting media, then carefully lower it over the drop so that the cover slip covers the medium without trapping air bubbles underneath. Use the corner of a paper towel to blot-up any excess DAPI mounting media at the edges of the cover slip.
- Set the coverslip down on DAPI mounting medium with the cells facing down. Make sure, the cells become dry if not, then it can be left at room temperature for more time. Then carefully seal with clear fingernail polish. Dry sealing in the dark at room temperature for 2 h.
Data analysis
Our protocol describes the visualization of macropinosomes in prostatic fibroblasts. These macropinosomes further fused into lysosome and get degraded by their acidic enzymes (lysozymes). To rule out the background signal possibility in labeling macropinosomes, we also included negative control group by treating Ras overexpressing mouse fibroblasts with macropinocytosis inhibitors. In addition to negative control, experimental methods were set up to observe macropinosomes fusion in lysosomes and its degradation by lysozymes. To track lysosomes, LysoTracker green and for degradation DQTM-BSA was used. The steps of the analysis are summarized below:
- We follow four criteria to identify macropinosomes:
- Dextran positive.
- Excluded from the nucleus.
- Surrounded by GFP-positive cytoplasm.
- Fluorescence intensities of macropinosomes are comparable with or higher than that of the extracellular space.
- We have had the best results imaging these samples using a Confocal Laser Scanning Microscope (Leica, Germany).
- Images were acquired with a 60x 1.4 NA oil immersion lens. Different lasers were used that span the blue (457, 477, and 488 nanometers), green (514 and 543 nanometers), and red (633 and 647 nanometers) spectral regions. Fluorescence was recorded in individual channels acquired in a sequential mode to avoid cross-talk using a highly sensitive 32-channel gallium arsenide phosphide detector. The pinhole was set to 1 Airy unit.
- As an example of the in vitro implementation of this protocol, we have assessed the extent of macropinocytosis in Ras-overexpressing mouse fibroblast. A representative example of a confocal image is shown in Figure 1.
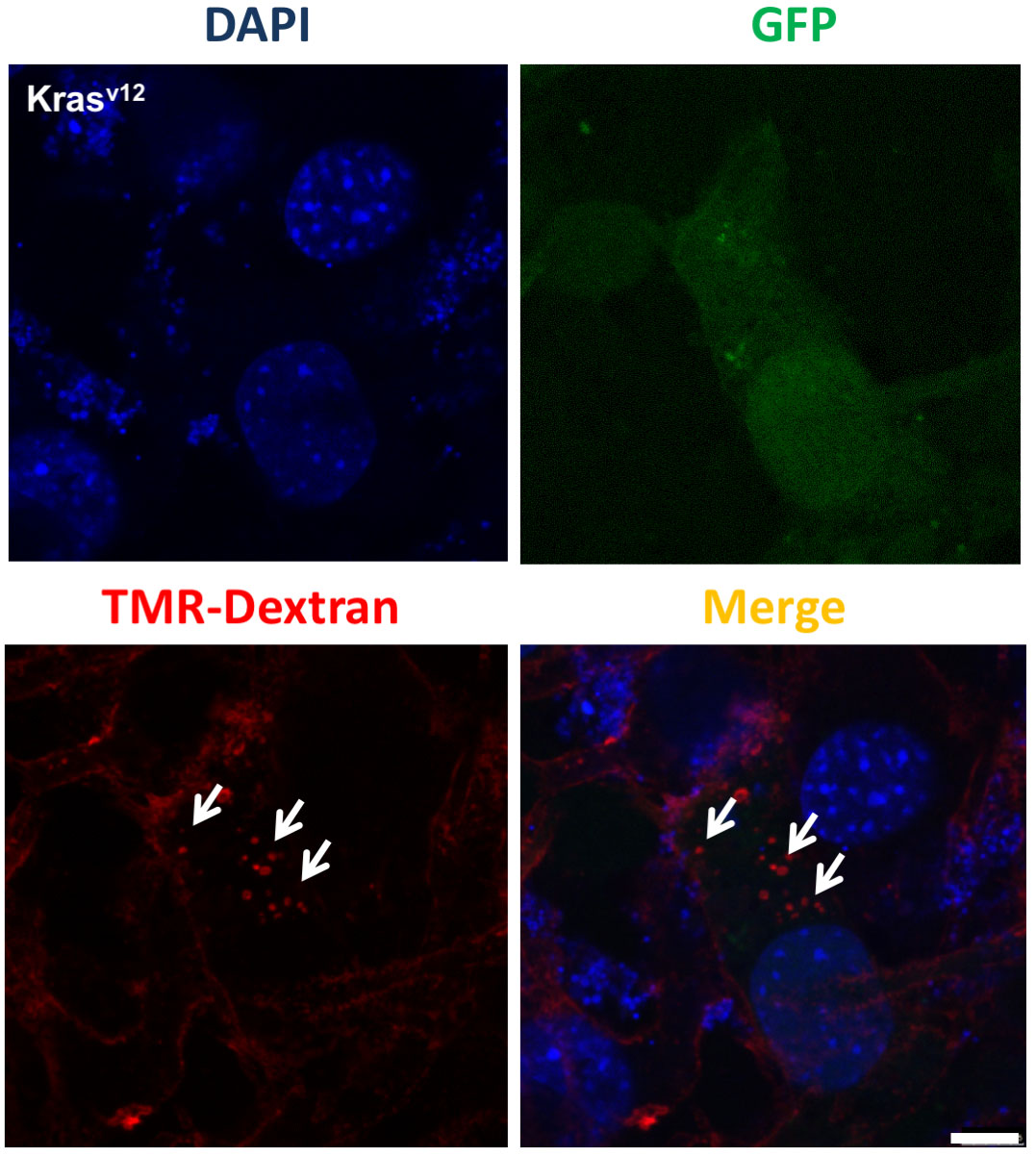
Figure 1. Monitoring of macropinosomes. Representative images show TMR-dextran–positive macropinosomes (arrowheads) in RasV12 prostatic mouse fibroblasts (expressing GFP). Scale bar = 7.5 μm. - Validation controls were analyzed as described below:
- Negative control: Grow the RasV12 mouse prostatic fibroblasts on coverslip as described in Procedure A. Treat cells with EIPA at 25 μM for 30 min prior to adding dextran in serum-free stromal media. EIPA is a clinical inhibitor of the Na+/H+ exchanger situated in the plasma membrane. Label the cells with TMR-dextran as described in Procedure B. Cells were fixed and analyzed by confocal microscopy. A representative example of a confocal image is shown in Figure 2.
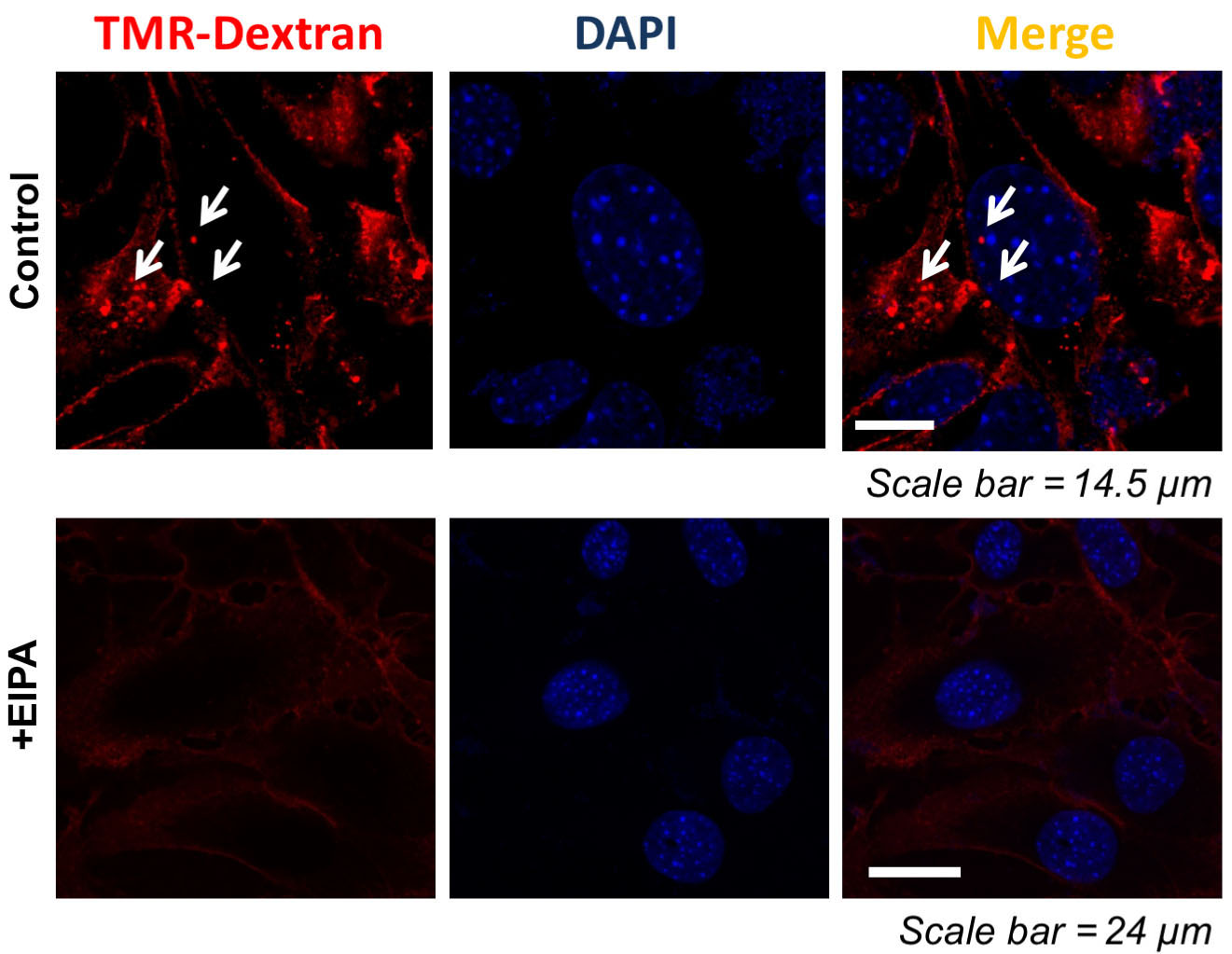
Figure 2. Reduction of macropinocytosis by EIPA. Macropinosomes visualization in RasV12 prostatic mouse fibroblasts was carried out with 30 min pretreatment with 5-(N-ethyl-N-isopropyl) amiloride (EIPA). Scale bar = 24 μm. - Monitoring of macropinosomes lysosome fusion: Grow the RasV12 mouse prostatic fibroblasts on coverslip as described in Procedure A. Cells are incubated with a mixture of Lyso tracker green DND-26 (75 nM) and TMR-dextran (1 mg/ml each) for 30 min at 37 °C. After 30 min, cells are placed on ice, washed for 10 min in ice-cold dye-free medium for three times, and fixed with 3.7% formaldehyde for 20 min on ice. The fluorescent signal emanating from Lysotracker with TMR-dextran–positive staining indicates lysosomal degradation of macropinosomes. Image was obtained using microscope (Figure 3).
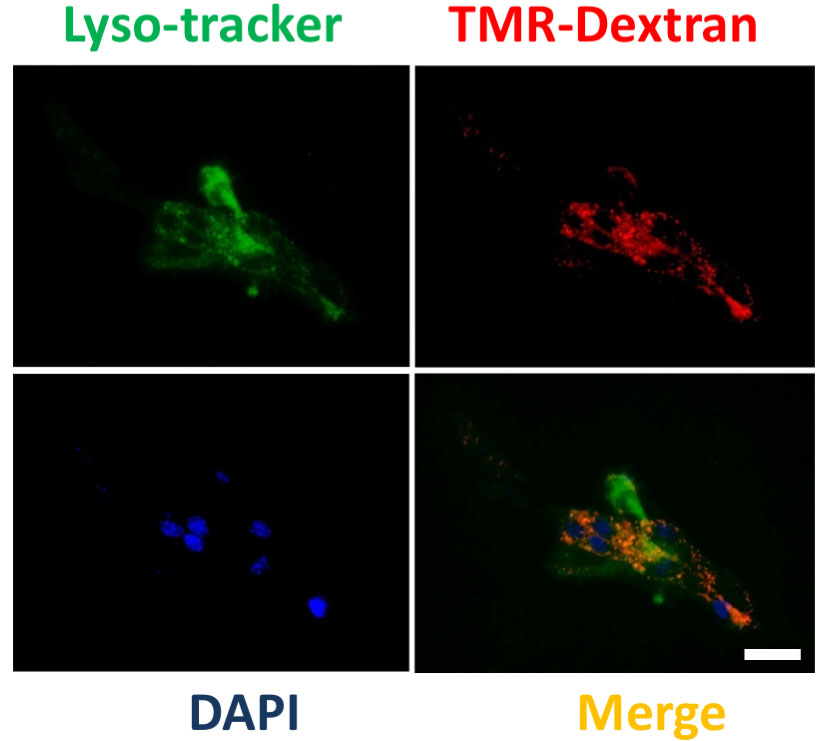
Figure 3. Monitoring internalization of macropinosomes in lysosomes. Acidification of macropinosomes was monitored by co-localization of LysoTracker (green) with TMR-Dextran suggesting fusion of macropinosomes with lysosomes (see orange puncta in the merged image). Images show representative microscopic images. Scale bar, 30 μm. - Test for albumin uptake by macropinosomes and lysosomal localization: Macropinocytosis substantially elevated uptake of proteins such as bovine serum albumin (BSA) from the extracellular fluid (Commisso et al., 2013). We choose DQTM-BSA to confirm albumin uptake and lysosomal function. DQTM-BSA is a quenched fluorogenic compound that requires enzymatic cleavage in an acidic intracellular compartment (i.e., lysosomes) for strong fluorescence. Grow the Cancer associated prostatic fibroblasts on coverslip as described in Procedure A. Cells are incubated with TMR-dextran and DQTM-BSA (10 μg/ml in serum free medium) for 1 h at 37 °C to assure macropinosomes formation and that the reagent reaches the lysosomal compartment. Cells were fixed and analyzed by confocal microscopy. A representative example of a confocal image is shown in Figure 4.
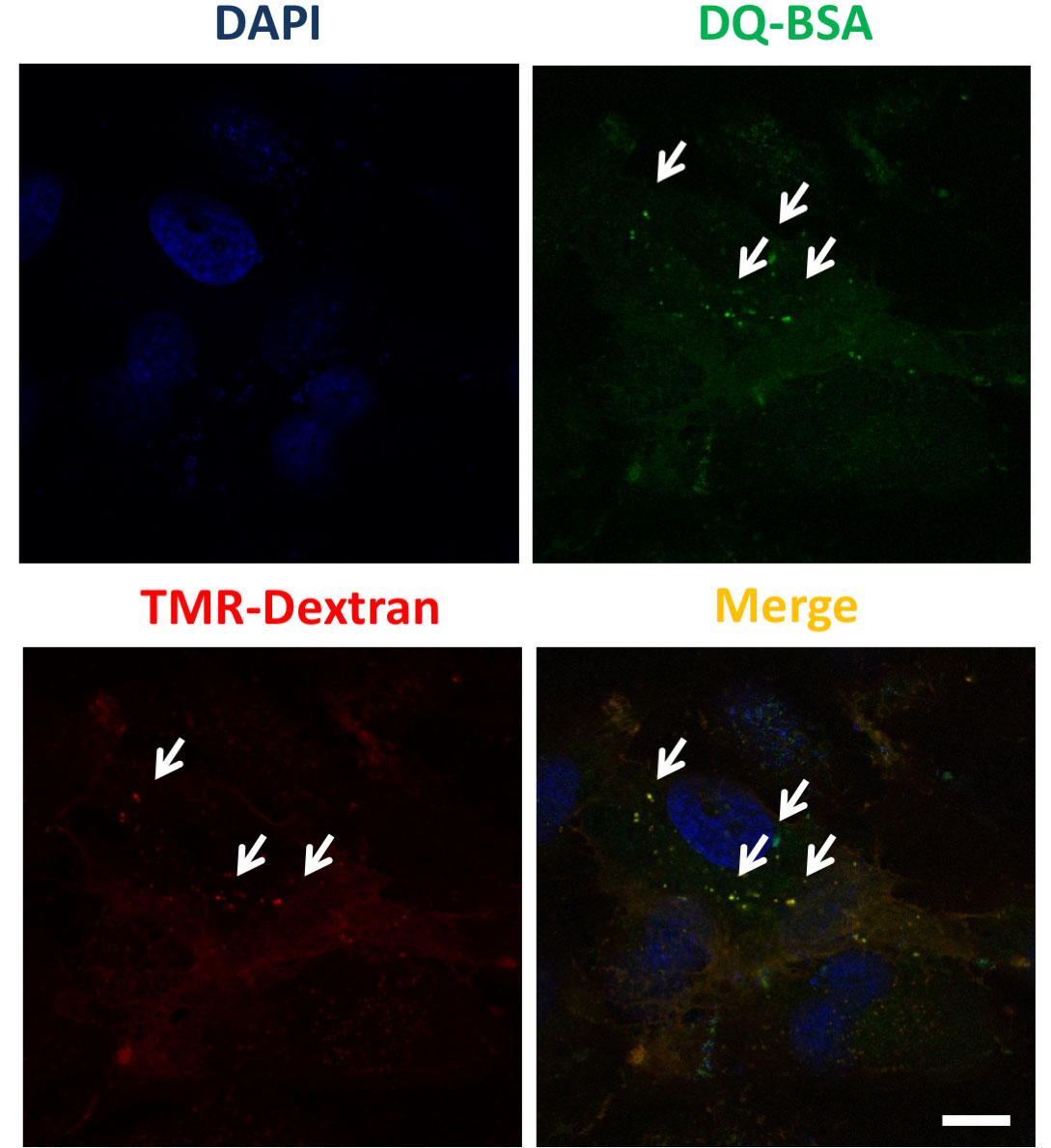
Figure 4. Monitoring of extracellular albumin internalization in macropinosomes. CAF that were co-incubated with fluorescent DQ-BSA (green) and TMR-dextran (red) then fixed after 1-h chase. The fluorescent signal emanating from DQ-BSA with TMR-dextran–positive staining (arrowheads) indicates albumin uptake by macropinosomes and subsequent lysosomal breakdown. Scale bar = 7.5 μm.
- Negative control: Grow the RasV12 mouse prostatic fibroblasts on coverslip as described in Procedure A. Treat cells with EIPA at 25 μM for 30 min prior to adding dextran in serum-free stromal media. EIPA is a clinical inhibitor of the Na+/H+ exchanger situated in the plasma membrane. Label the cells with TMR-dextran as described in Procedure B. Cells were fixed and analyzed by confocal microscopy. A representative example of a confocal image is shown in Figure 2.
Recipes
- Stromal complete medium
DMEM/F12 media
5% FBS
5% NuSerum (HyClone)
0.1% Insulin - PBS 1x, pH 7.4 (1 L)
NaCl 9 g
NaH2PO4 0.23 g
Na2HPO4 1.15 g
Sterile distilled water, up to 1 L - Dextran stock solution
Dissolve 25 mg of fluorescently labeled 70-kDa dextran in 1.25 ml of PBS to obtain a final concentration of 20 mg/ml dextran stock solution
Store stock solution in 100-μl aliquots in the dark at -20 °C
Note: Make sure that dextran solution becomes clear and transparent. Dextran takes 10-15 min to dissolve completely by pipetting up and down. - EIPA stock solution
EIPA stock solutions (10 mM) are prepared in Dimethyl sulfoxide (DMSO) and used at a working concentration of 25 mM - LysoTracker green DND-26 Green
- Allow a vial of 1 mM LysoTracker DND-26 solution (50 μl in DMSO) (Invitrogen L7526) to warm to room temperature
- Briefly centrifuge to deposit the solution at the bottom of the vial before opening
- From the 1 mM solution, prepare a 500 nM stock solution using molecular grade H2O
- Cover in aluminum foil to minimize light exposure
- Store stock solution below -20 °C
- Immediately before use, prepare a staining solution of roughly 75 nM LysoTracker Green by combining 1.5 μl of the 500 nM stock solution with 8 μl of molecular-grade H2O
- DQ-Green BSA stock solution
- Dissolve 1 mg of lyophilized powder of DQ-BSA in 500 μl of PBS to make a stock of 2 mg/ml of DQ-Green BSA
- Mix thoroughly by vortexing and pipetting
- After reconstitution wrap it with aluminum foil to protect it from light and store at 4 °C
- Fixation buffer
Add 37% (vol/vol) ACS reagent-grade formaldehyde solution to PBS to a final concentration of 3.7% (vol/vol)
Note: Use freshly prepared solution.
Acknowledgments
This work was supported by grants from the National Cancer Institute (CA108646 to NAB) and Veterans Affairs (BX001040 to NAB). Support by Enhanced Seed grants EF/2018-19/QE04-11 (to R.M.) from Manipal University Jaipur, Rajasthan, India is gratefully acknowledged.
Competing interests
The authors declare that they have no conflict of interest.
Ethics
In accordance with institutional animal care and use committee approval, primary mouse fibroblasts were harvested and grown using approved procedures.
References
- Bar-Sagi, D. and Feramisco, J. R. (1986). Induction of membrane ruffling and fluid-phase pinocytosis in quiescent fibroblasts by ras proteins. Science 233(4768): 1061-1068.
- Commisso, C., Davidson, S. M., Soydaner-Azeloglu, R. G., Parker, S. J., Kamphorst, J. J., Hackett, S., Grabocka, E., Nofal, M., Drebin, J. A., Thompson, C. B., Rabinowitz, J. D., Metallo, C. M., Vander Heiden, M. G. and Bar-Sagi, D. (2013). Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature 497(7451): 633-637.
- Davidson, S. M., Jonas, O., Keibler, M. A., Hou, H. W., Luengo, A., Mayers, J. R., Wyckoff, J., Del Rosario, A. M., Whitman, M., Chin, C. R., Condon, K. J., Lammers, A., Kellersberger, K. A., Stall, B. K., Stephanopoulos, G., Bar-Sagi, D., Han, J., Rabinowitz, J. D., Cima, M. J., Langer, R. and Vander Heiden, M. G. (2017). Direct evidence for cancer-cell-autonomous extracellular protein catabolism in pancreatic tumors. Nat Med 23(2): 235-241.
- Lim, J. P. and Gleeson, P. A. (2011). Macropinocytosis: an endocytic pathway for internalising large gulps. Immunol Cell Biol 89(8): 836-843.
- Mishra, R., Haldar, S., Placencio, V., Madhav, A., Rohena-Rivera, K., Agarwal, P., Duong, F., Angara, B., Tripathi, M., Liu, Z., Gottlieb, R. A., Wagner, S., Posadas, E. M. and Bhowmick, N. A. (2018). Stromal epigenetic alterations drive metabolic and neuroendocrine prostate cancer reprogramming. J Clin Invest 128(10): 4472-4484.
- Swanson, J. A. and Watts, C. (1995). Macropinocytosis. Trends Cell Biol 5(11): 424-428.
Article Information
Copyright
© 2019 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Mishra, R. and Bhowmick, N. A. (2019). Visualization of Macropinocytosis in Prostate Fibroblasts. Bio-protocol 9(10): e3235. DOI: 10.21769/BioProtoc.3235.
Category
Cancer Biology > Cancer biochemistry > Drug resistance
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









