- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
3D Co-culture System of Mouse Prostatic Wild-type Fibroblasts with Human Prostate Cancer Epithelial Cells
Published: Vol 9, Iss 9, May 5, 2019 DOI: 10.21769/BioProtoc.3225 Views: 5871
Reviewed by: Gal HaimovichMarzia Di DonatoSalma Merchant

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Capturing the Driving Role of Tumor-host Crosstalk in a Dynamical Model of Tumor Growth
Sebastien Benzekry [...] Lynn Hlatky
Nov 5, 2015 9330 Views

A Simplified 3D-Plasma Culture Method for Generating Minimally Manipulated Autologous Equine Muscle-Derived Progenitor Cells
Hélène Graide [...] Didier Serteyn
Dec 5, 2025 1251 Views

Simple and Rapid Model to Generate Differentiated Endometrial Floating Organoids
Adriana Bajetto [...] Tullio Florio
Feb 5, 2026 93 Views
Abstract
Heterogeneous prostatic carcinoma-associated fibroblasts (CAF) contribute to tumor progression. This was established using transgenic mouse models. Paracrine interactions between fibroblasts and epithelial cells were further interrogated using isolated 2D cell culture systems, but 3D culture systems currently being developed can better mimic reciprocal interactions potentially found in the native tissue. To understand paracrine and juxtacrine signaling among fibroblasts and epithelia, 3D co-cultures with species differences allows for further subsequent analysis of the cultures. The use of mouse and human cells, for example, in one system allows for species-specific FACS or quantitative PCR analysis. This protocol describes the use of a 3D Co-culture System of Mouse Prostatic Wild-type Fibroblasts with Human Prostate Cancer Epithelial Cells.
Keywords: 3D Co-cultureBackground
Prostate cancer (PCa) is a heterogeneous disease that results in the second highest cancer mortality in men. From the early steps of PCa initiation, the associated stromal fibroblastic cells begin to co-evolve with cancer progression and are predictive of recurrent disease and survival (Ayala et al., 2003). Studying paracrine signaling from the epithelial cancer cells to the associated fibroblasts and vice versa is imperative to understand the complex interactions during all stages of cancer progression (Bhowmick et al., 2004; Placencio et al., 2008). 2D culture systems where conditioned media from one cell type is incubated with the target cell type as well as those utilizing Boyden chamber cultures enable the exchange of paracrine factors. However, the characteristics of the cells in collagen matrix have documented differences in their responses to hormones and growth factors (Ziaee and Chung, 2014; Naba et al., 2014). Utilizing a 3D co-culture system allows a more representative readout of paracrine interactions between the stroma and epithelia. This system can be adapted for multiple combinations of stromal fibroblasts and cancer epithelial cells. Combinations of mouse and human cells can be used to better differentiate the cell type from which the signals were derived/received. Furthermore, incubation in a hypoxic chamber can more closely model physiologic environments. We have used localized prostate cancer as an example for our studies.
Materials and Reagents
- Pipette tips
- Sterile 1.5 ml microcentrifuge tubes
- Sterile 15 ml conical tubes
- 6-well cell culture plate (Fisher Scientific, catalog number: 08-772-49)
- Flat bottom 96-well plate (Fisher Scientific, catalog number: 07-200-90)
- 10 cm Tissue culture dish (Fisher Scientific, catalog number: 08-772E)
- Nylon mesh squares cut 2 cm x 2 cm (Elko Filtering Co., catalog number: 03-80/37)
- Metal grids, cut 2 cm x 2 cm, sterlized (Charcoal aluminum screen patches, Phifer incorporated, 3025123)
- FACS tubes (Corning, catalog number: 352235)
- Primary cultured mouse wild type prostatic fibroblasts (generated as previously described by Kato et al., 2018; Mishra et al., 2018)
- Human CWR22Rv1 epithelial cells (ATCC, catalog number: CRL-2505)
- 10x DMEM medium (Fisher Scientific, catalog number: SH3000203)
- Trypsin (Fisher Scientific, catalog number: MT25053CI)
- RPMI medium (Fisher Scientific, catalog number: SH3001103)
- DME/F12 medium (Fisher Scientific, catalog number: SH300404)
- Fetal Bovine Serum (Atlanta Biologicals, catalog number: S11550)
- NuSerum (Fisher Scientific, catalog number: 355500)
- Collagenase Type I (Invitrogen, catalog number: 17100-017)
- Dispase (Invitrogen, catalog number: 17105-041)
- BSA (MP Biomedicals, catalog number: 810034)
- Glutaraldehyde, 25% solution (Sigma, catalog number: G6257)
- Gentamicin (Life Technologies, catalog number: 15710-064)
- Insulin (human recombinant zinc, Gibco Life Technologies, catalog number: 12585-014)
- Testosterone (Testosterone propionate C-IIIN from Sigma-Aldrich, catalog number: T1875-5g)
- Anti-human CD105-APC antibody (eBiosciences, catalog number: 17-1057-41)
- Anti-human EpCAM-PE antibody (eBiosciences, catalog number: 12-9326-41)
- Rat tail collagen I (generated as previously described [Kato et al., 2018; Mishra et al., 2018], 6 month shelf life at 4 °C)
- Matrigel (BD Biosciences, catalog number: 354234)
- EBSS, 10x (Sigma-Aldrich, catalog number: E7510)
- NaHCO3
- NaOH
- Sterile distilled water
- NaCl
- NaH2PO4
- Na2HPO4
- RPMI complete epithelial medium (for CWR22Rv1) (see Recipes)
- DME/F12 Stromal complete medium (for mouse WT fibroblasts) (see Recipes)
- Setting solution (see Recipes)
- 1x PBS (See Recipes)
- Digestion solution (see Recipes)
Equipment
- Sterile forceps (Fine Science Tools, catalog number: 11254-20)
- Sterile Scissors (Fine Science Tools, catalog number: 14060-09)
- Sterile 10 ml serological pipette
- Biosafety cabinet for tissue culture work
- Pipette-aid
- Centrifuge with adaptors for 15-ml conical tubes, for use at room temperature
- Humidified cell culture incubator set to 37 °C and 5% CO2
- Humidified cell culture incubator set to 37 °C, 5% CO2, and 2% O2
- BD LSRII for flow cytometry
- Centrifuge for Eppendorf tubes
Software
- FlowJo software v10.3 was used for FACS analysis
Procedure
Day 1
Note: Step one may be performed the day before, keep sterile.
- Preparation of the collagen-coated nylon (For use in Day 2 Step 1, you will need 100-200 µl per nylon)
For 1 ml (keep on ice):
Collagen, Type I 70% (700 µl) (Range of concentration is between 1 mg/ml to 10 mg/ml)
10x DMEM 10% (100 µl)
1x DME/F12 stromal complete medium 5% (50 µl)
1x complete epithelial RPMI media (50 µl)
FBS 5% (50 µl)
NuSerum 5% (50 µl)
Setting solution (usually 25%-35% of collagen amount for a pink-red color similar to medium) - Autoclave Nylon mesh squares ~2 cm x 2 cm, using dry program. Autoclave all squares in any autoclavable container that can be accessed with forceps to remove nylons one at a time.
- Place autoclaved Nylon mesh squares on a sterile tissue culture plate, 10 cm.
- Coat the nylon squares with the collagen matrix (100-200 µl for each) using a sterile pipette, see Figure 1, top row.
- Incubate at 37 °C for 30 min, 5% CO2.
- Add 1% glutaraldehyde (diluted in sterile H2O) to submerge the nylon squares (use about 10 ml total volume for one 10 cm plate), incubate at 4 °C 1 h.
- After fixation, wash with sterile 1x PBS 4 times, and wash with 1x DME/F12 1 time, and add 1x DME/F12 until use, store at 4 °C.
- Trypsinize cells*
Depending on the total number of gel plugs desired, at least one 10 cm plate of each cell type is recommended and will vary for each cell type. Sub-confluence is recommended as actively proliferating cells are best. Cells will be trypsinized and resuspended in either 1x complete RPMI or DME/F12 media.
Note: It is best to use cells that are actively proliferating and not confluent. - Count cells
Wash cell pellet twice in 1x PBS, keep on ice. Each gel plug has 6 x 105 stromal fibroblastic cells + 2 x 105 epithelial cells. Make a single master mix of epithelial with fibroblastic cells, and pellet just before adding the matrix. Prepare 1 extra gel’s worth of cells since some volume will be lost with pipetting the plugs. - Preparation of the matrix
Note: Keep on ice, *This step can be started in parallel with the cell trypsinization steps (*Step 4) during wait times of trypsinization or wash steps.
For 1 ml:
Collagen 50% (500 µl) (Range of concentration is between 1 mg/ml to 10 mg/ml)
Matrigel 20% (200 µl)
10x DMEM 10% (100 µl)
1x DME/F12 stromal complete medium 5% (50 µl)
1x complete epithelial RPMI media 5% (50 µl)
FBS 5% (50 µl)
NuSerum 5% (50 µl)
Setting solution is added after all other ingredients and will increase the total volume of matrix over 1 ml (Add 25%-35% of the collagen amount (125 μl to 175 μl depending on collagen starting concentration, more concentrated collagen may require more setting solution) to make a pink-red color). This will allow the matrix to reach the proper pH to keep cells viable and solidify. - Add matrix into aspirated cell pellet, mix well and avoid making bubbles.
- Seeding cells in gel plugs: Pipette 150 µl cell-matrix mix in a flat bottom 96-well dish (it’s recommended to plate mix in outer edges of plate, incubate at 37 °C for 30 min (no more than 60 min to avoid drying out). Recommend plating 3 gel plugs per treatment condition.
- When gel plugs have solidified after 30 min, add 100 μl media per well (RPMI complete medium as stated in Recipes + DME/F12 complete medium as stated in Recipes, equal volumes of each medium). Return the plate to 37 °C to incubate overnight.
- Assembly of the 3D system. In a 6-well plate assemble the 3D system. In each well, using sterile forceps, place 1 autoclaved metal grid cut slightly larger than 2 cm x 2 cm (and bent to stand like a table, see Figure 1, second row) carefully upright in well, grids may slide around, so try to position in the center of well. Then center collagen coated nylon on top.
- Pop out cell-matrix gel plugs prepared on the previous day with the cell mixture (see Figure 1, bottom row) by pipetting 200 µl medium against the side of each well, using a 200 µl pipette. Catch gels in a sterile 10 cm dish containing medium as they slide out of the wells.
- Placing gel plugs on grid. Transfer 3 gels onto each collagen coated nylon with sterile forceps and around add 2-3 ml medium to just touch the bottom of the nylon to allow wicking to each of the individual gel plugs. Do not immerse the gels completely in the media, they must have the top surface exposed to the air. Treatments may be added to the media and changed as often as desired.
- Replenish medium every other day or as needed if the medium level has dropped below nylon and culture up to 10 days in hypoxic conditions (2% O2).
Notes:- The limit to keep gel plugs has been 10 days in our hands. Active cells start digesting the matrix and the gel plug form is slowly dissolved away, likely from cells digesting and degrading the matrix components. Therefore, the 3D structure is diminished by 10 days and we feel this is not comparable to the 3D structures in earlier time points. We typically use time points between 24 and 72 h, and sometimes up to 5 days.
- For experiments involving CD105, hypoxia is needed as it better mimics the in vivo conditions in the body. Other readouts may not need hypoxia.
End of Experiment
- Cell harvesting: When experiment is finished, prepare fresh digestion solution.
- Place gel into Eppendorf tubes with 500 µl digestion solution. Cut gels with scissors into small pieces, incubate at 37 °C for 15 min. Shake tubes once by hand during incubation (or keep on rotation device for continuous gentle mixing; vortexing is not recommended). If gels are not dissolved after 15 min, incubate for another 5 min.
- Spin to pellet cells, at 300 x g for 2 min.
- FACS Analysis: Stain cells using any standard FACS protocol such as the one provided below, keep cells on ice.
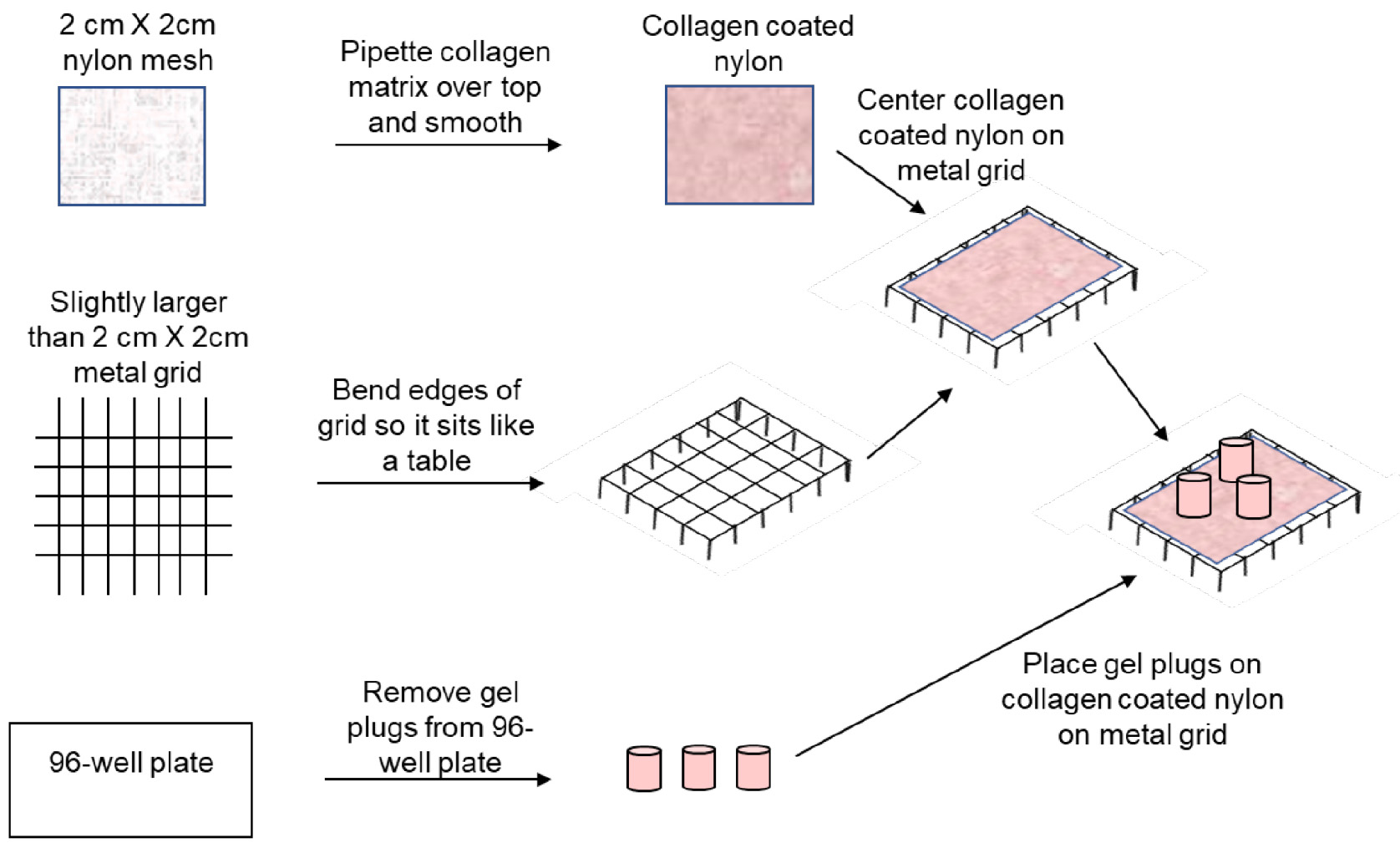
Figure 1. 3D Co-Culture System Assemby. Using sterile conditions, the nylon mesh was coated in the collagen matrix, allowed to solidify, crosslinked with glutaraldehyde, and was washed as described. Metal grids were bent to form tables that fit into a 6-well plate. The collagen coated nylon was centered on each metal grid table. Gel plugs were removed from a 96-well plate and may be kept in media to prevent drying. Gel plugs were then transferred to the top of the collagen-coated nylon as shown. Media was then added to each well to touch the collagen-coated nylon without submerging the gel plugs completely.
FACS Extracellular staining
- Wash cells with 500 µl PBS + 1% BSA, repeat once.
- After last wash, aspirate solution and leave 100 µl in the tube with the pelleted cells.
- Add extracellular staining antibodies (1 µl antibody for 1 x 106 cells in 100 µl PBS + 1% BSA), swirl to mix cells and incubate in the dark on ice for 20-30 min.
- Spin at 300 x g for 2-5 min and wash with 200 µl PBS + 1% BSA, repeat.
- Transfer cells to FACS tubes with ~400 µl PBS + 1% BSA, keep on ice.
- Run cells on BD LSR II.
Data analysis
- Import files into FlowJo software.
- Gate cells to include live cells for analysis. Exclude cells in the lower left corner.
- Gate cells for EpCAM+ (epithelial cells) versus EpCAM-.
- EpCAM+ cells were gated for CD105+. EpCAM+/CD105+ cells are the epithelial cell population with CD105 expression.
Note: The use of other FACS antibodies may be used to analyze the expression of other target proteins of interest. - The percentage of these cells can be used for statistical analysis. At least three replicates are recommended to calculate standard deviation. Examples can be found in Kato et al., 2018.
Notes
- When working with collagen or Matrigel, keep on ice as much as possible to prevent solidifying.
- It is best to use cells that are actively proliferating and not confluent.
- Keep all reagents and materials sterile.
- This 3D protocol has been used to obtain digested cells for RNA analysis using species-specific primers to differentiate epithelial versus stromal cell expression of desired genes.
- The limit to keep gel plugs has been 10 days in our hands. Active cells start digesting the matrix and the gel plug form is slowly dissolved away, likely from cells digesting and degrading the matrix components. Therefore, the 3D structure is diminished by 10 days and we feel this is not comparable to the 3D structures in earlier time points. We typically use time points between 24 and 72 h, and sometimes up to 5 days.
- Cells are expected to double every 48 h or as indicated for specific cell types.
- DME/F12 stromal was used for our primary fibroblasts as this is what they are usually grown in, but other stromal cell types may be substituted with their preferred stromal media.
- For the experimental readout using EpCAM+ cells, if the epithelial cells are thought to undergo epithelial to mesenchymal transition, EpCAM+ staining may not fully capture the epithelial cell population and alternative markers would be necessary (such as the use of stably transduced fluorescent cells).
Recipes
- RPMI complete epithelial medium (for CWR22Rv1)
RPMI media
10% FBS
0.1% Gentamicin antibiotic - DME/F12 Stromal complete medium (for mouse WT fibroblasts)
DME/F12 media
5% FBS
5% NuSerum
0.1% Insulin
0.1% Gentamicin antibiotic
Add fresh testosterone to the media (10-8 M) each time it’s replenished - Setting solution
Mix together until dissolved, filter sterilize with a 0.20 µm syringe filter, aliquot and store at -20 °C10x EBSS 100 ml NaHCO3 2.45 g 1 M NaOH 7.5 ml Sterile distilled water 42.5 ml - 1x PBS, pH 7.4 (1 L)
Sterile distilled water, up to 1 LNaCl 9 g NaH2PO4 0.23 g Na2HPO4 1.15 g - 500 µl digestion solution (for each gel plug)
Collagenase (20 mg/ml) 125 μl Dispase (10 mg/ml) 167 μl RPMI complete Media 208 μl
Acknowledgments
This work was supported by grants from the National Cancer Institute (CA108646 to NAB and CA098912 to VRP) and Veterans Affairs (BX001040 to NAB).
Competing interests
The authors declare that they have no conflict of interest.
Ethics
In accordance with institutional animal care and use committee approval, primary mouse fibroblasts were harvested and grown using approved procedures.
References
- Ayala, G., Tuxhorn, J. A., Wheeler, T. M., Frolov, A., Scardino, P. T., Ohori, M., Wheeler, M., Spitler, J. and Rowley, D. R. (2003). Reactive stroma as a predictor of biochemical-free recurrence in prostate cancer. Clin Cancer Res 9(13): 4792-4801.
- Bhowmick, N. A., Chytil, A., Plieth, D., Gorska, A. E., Dumont, N., Shappell, S., Washington, M. K., Neilson, E. G. and Moses, H. L. (2004). TGF-beta signaling in fibroblasts modulates the oncogenic potential of adjacent epithelia. Science 303(5659): 848-851.
- Kato, M., Placencio-Hickok, V. R., Madhav, A., Haldar, S., Tripathi, M., Billet, S., Mishra, R., Smith, B., Rohena-Rivera, K., Agarwal, P., Duong, F., Angara, B., Hickok, D., Liu, Z. and Bhowmick, N. A. (2018). Heterogeneous cancer-associated fibroblast population potentiates neuroendocrine differentiation and castrate resistance in a CD105-dependent manner. Oncogene 38: 716-730.
- Mishra, R., Haldar, S., Placencio, V., Madhav, A., Rohena-Rivera, K., Agarwal, P., Duong, F., Angara, B., Tripathi, M., Liu, Z., Gottlieb, R. A., Wagner, S., Posadas, E. M. and Bhowmick, N. A. (2018). Stromal epigenetic alterations drive metabolic and neuroendocrine prostate cancer reprogramming. J Clin Invest 128(10): 4472-4484.
- Naba, A., Clauser, K. R., Lamar, J. M., Carr, S. A. and Hynes, R. O. (2014). Extracellular matrix signatures of human mammary carcinoma identify novel metastasis promoters. Elife 3: e01308.
- Placencio, V. R., Sharif-Afshar, A. R., Li, X., Huang, H., Uwamariya, C., Neilson, E. G., Shen, M. M., Matusik, R. J., Hayward, S. W. and Bhowmick, N. A. (2008). Stromal transforming growth factor-beta signaling mediates prostatic response to androgen ablation by paracrine Wnt activity. Cancer Res 68(12): 4709-4718.
- Ziaee, S. and Chung, L. W. (2014). Induction of integrin α2 in a highly bone metastatic human prostate cancer cell line: roles of RANKL and AR under three-dimensional suspension culture. Mol Cancer 13: 208.
Article Information
Copyright
© 2019 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Placencio-Hickok, V. R., Kato, M. and Bhowmick, N. A. (2019). 3D Co-culture System of Mouse Prostatic Wild-type Fibroblasts with Human Prostate Cancer Epithelial Cells. Bio-protocol 9(9): e3225. DOI: 10.21769/BioProtoc.3225.
Category
Cancer Biology > Oncogenesis > Ex vivo tissue culture model
Cancer Biology > Proliferative signaling > Tumor microenvironment > Proliferation analysis
Cell Biology > Cell isolation and culture > 3D cell culture
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









