- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Separation of Natural Collagen Crosslinks Using Buffer and Ion-pairing Agent Free Solvents on Silica Hydride Column for Mass Spectrometry Detection
Published: Vol 9, Iss 9, May 5, 2019 DOI: 10.21769/BioProtoc.3224 Views: 4762
Reviewed by: Roopali AgarwalVasudevan AchuthanAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Protein Turnover Dynamics Analysis With Subcellular Spatial Resolution
Lorena Alamillo [...] Edward Lau
Aug 5, 2025 2397 Views

Immunopeptidomics Workflow for Isolation and LC-MS/MS Analysis of MHC Class I-Bound Peptides Under Hypoxic Conditions
Hala Estephan [...] Eleni Adamopoulou
Nov 20, 2025 1848 Views

Quantitative Proteomics of Nitrosylated Proteins in Melanoma Using the Biotin-Switch Technique Combined With Tandem Mass Tag Labeling
Vipin K. Yadav [...] Sanjay Premi
Dec 5, 2025 1540 Views
Abstract
In this protocol we describe the separation of collagen crosslinks in biological tissues and samples including skin, tendon, cartilage, bone and urine. The existing methods use either cation exchange chromatography followed by post-column derivatization with ninhydrin or reverse phase chromatography with mass spectrometry detection. The cation exchange chromatography method has limited sensitivity and long run times while reverse phase chromatography requires strong ion-pairing. In this method, the sample containing crosslinks is applied on a diamond hydride column using water and acetonitrile solvents containing 0.1% (w/v) formic acid. Eight crosslinks are eluted separately from the column and detected by mass spectrometry in the sub-pmol range. By using this method, it is possible to separate all crosslinks of collagen in several biological samples without the need for ion-pairing agent or derivatization for detection.
Keywords: SkinBackground
Collagen is the major structural protein and a major component of skin, bone, cartilage, tendon and dentin in teeth. Collagen structure is stabilized by several crosslinks which are found in small amounts in collagen where they are present in different types and varying ratios, depending on the tissue type. Alteration in crosslink types and amount results in several diseases such as bone fragility and skin hyperelasticity (Pinnell et al., 1972 and Arseni et al., 2018).
The development of a sensitive quantitative method using Liquid Chromatography-Mass Spectrometry (LC-MS) is essential for measuring the collagen crosslinks in healthy and diseased tissues. The existing methods for the quantitation of collagen crosslinks including cation exchange chromatography and reverse phase column require sodium buffers and strong ion pairing agents respectively (Avery et al., 2009 and Gineyts et al., 2010). These requirements make them unsuitable for subsequent analysis using Electrospray Ionization Mass Spectrometry (ESI-MS) where high buffer concentrations and ion pairing agents are known to suppress the mass spectrometry signal resulting in significant loss of sensitivity. Also Histidinohydroxylysinonorleucine (HHL) and Histidinohydroxymerodesmosine (HHMD) crosslinks have not previously been detected by mass spectrometry.
We developed a quantitative method for collagen crosslinks on silica hydride column that does not require buffer or ion-pairing agent which allows the use of mass spectrometry (LC-MS) for detection with high sensitivity. Additionally, this method separates and quantitates eight crosslinks in collagenous tissue quicker (total run time less than 10 min) than those of all previously reported methods. This method allows the detection of collagen crosslinks in a complex mixture using MS without the need for buffers or ion-pairing agents.
Materials and Reagents
- Pipette tips
- 1-10 μl (Thermo Fisher, FinntipTM, catalog number: FNP4641040N)
- 10-100 μl (Thermo Fisher, FinntipTM, catalog number: FNP4641070N)
- 100-1,000 μl (Thermo Fisher, FinntipTM, catalog number: FNP4641100N)
- Desmosine (DES) (Sigma-Aldrich, catalog number: D9439)
- Dihydroxylysinonorleucine (DHLNL) (Santa Cruz Biotechnology, catalog number: sc-207059)
- Lysinonorleucine (LNL) (TRC-Canada, catalog number: L488750)
- Pyridinoline (PYR) (BOC Science, catalog number: 77464-35-8)
- Deoxypyridinoline (DPYR) (BOC Science, catalog number: 83462-55-9)
- Hydroxylysinonorleucine (HLNL) (purified in our laboratory, see Naffa et al., 2019a)
- Histidinohydroxylysinonorleucine (HHL) (purified in our laboratory, see Naffa et al., 2019a)
- Histidinohydroxymerodesmosine (HHMD) (purified in our laboratory, see Naffa et al., 2019a)
- Mass spectrometry grade water (Fisher Chemicals, catalog number: FSBW5-4)
- Mass spectrometry grade acetonitrile (Fisher Chemicals, catalog number: FSBA955-4)
- Mass spectrometry grade formic acid (Fisher Chemicals, catalog number: FSBA117-50)
- Acetic acid (UNIVAR, catalog number: UN2789)
- Solvent A (see Recipes)
- Solvent B (see Recipes)
Equipment
- Autosampler amber vials (32 mm x 11.6 mm) (Thermo Fisher, catalog number: THC11090519)
- Vortex (Fisher Scientific, catalog number: FB15012)
- Automated finnpipettes (Thermo Fisher, catalog number: FNP4700850N)
- Thermo Fisher UHPLC with a Q-Exactive Orbitrap Mass Spectrometer equipped with a heated electrospray ionization (HESI) (Thermo Fisher, CA, USA)
Note: Table 1 shows all the specifications of the instrumentation in Equipment 4.
Table 1. Specifications of the Q-Exactive mass spectrometer UHPLC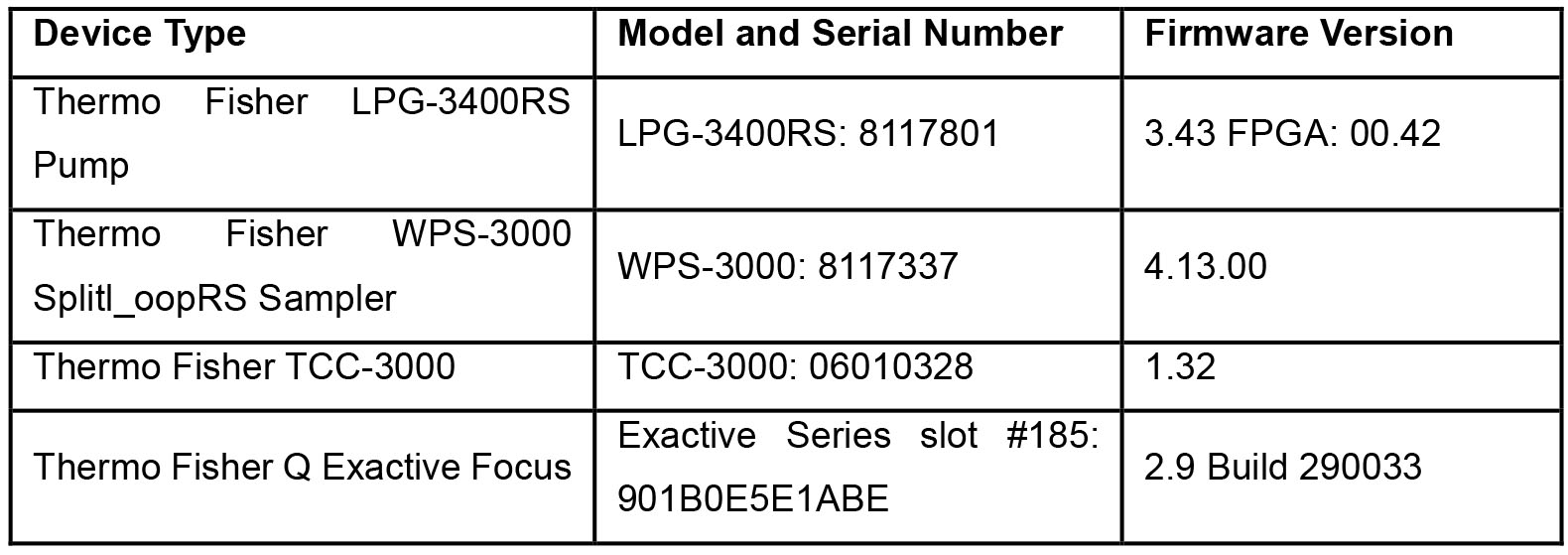
- Cogent Diamond Hydride HPLC column 150 mm x 2.1 mm; particle size, 2.2 µm; pore size 100 Å (Microsolv Technology, catalog number: 70200-15P-2)
Software
- Thermo Xcalibur 4.1.31.9 June 8, 2017 for qualitative analysis and quantitation analysis (Thermo Fisher, CA, USA)
Procedure
- Prepare the crosslink standards by serial dilution as shown in Table 2. Use water containing 0.1% formic acid as diluent and measure all volumes in μl and transfer them into amber autosampler vials.
Notes:- Different volumes can be used but you must use the same dilution factor.
- The prepared crosslink standard can be stored at -20 °C for up to one year, however, always make one new standard to check the concentration of the frozen ones.
- PYR and DPYR may degrade over time and their concentrations change by exposure to light, therefore always use amber autosampler vials and store them in the dark.
Table 2. Standard concentrations in µM of crosslinks diluted by water containing 0.1% formic acid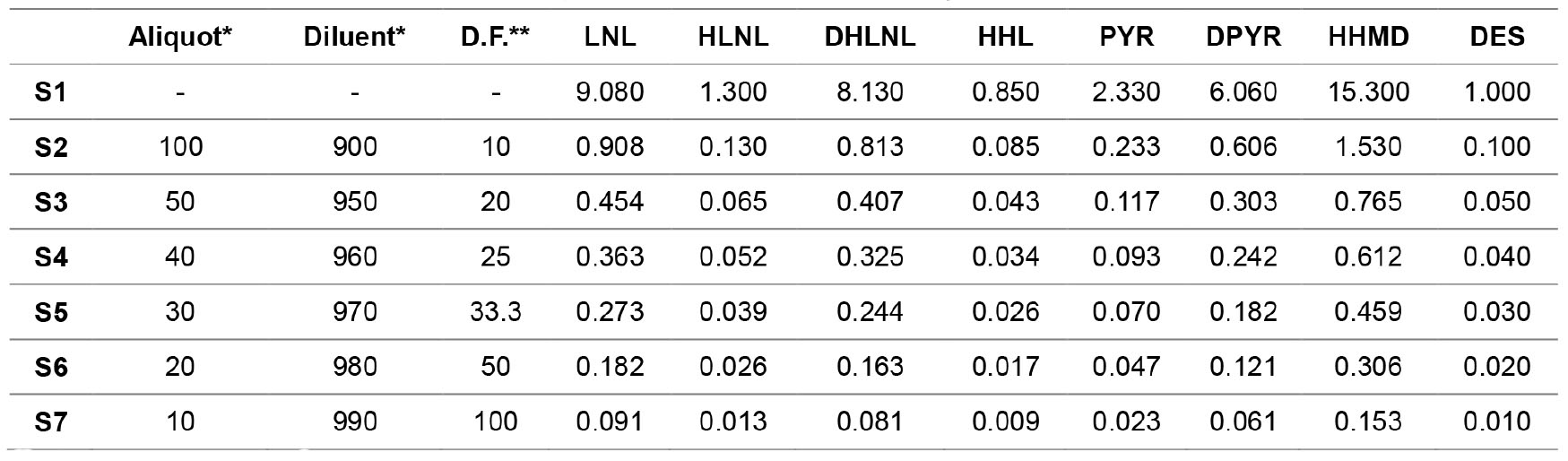
*Volume measured in μl.
**D.F. is dilution factor. - Set the temperature of the autosampler at 6 °C then place the vials in the autosampler.
- Use the following separation procedure on diamond hydride column:
- Flow rate of 0.3 ml/min.
Note: Higher flow rates can be also used (0.4-0.6 ml/min) which will elute the crosslinks faster, however this may affect the resolution. - Column temperature 25 °C.
- Flow rate of 0.3 ml/min.
- Use the following parameter for the mass spectrometry:
- Positive ion mode.
- Mass scans (100-700 m/z).
- Spray voltage 3,300 V.
- The capillary 320 °C.
- The probe heater temperatures 350 °C.
- Nitrogen gas set at flow rate of 10 L/min.
- Set the injection volume at 1.0 μl.
Note: Injected volume can be adjusted up to 10.0 μl depending on the crosslink concentration in the sample. - Solvent A is water and solvent B is 80% acetonitrile in water both containing 0.1% (v/v) formic acid (see Recipes).
- Use gradient elution program as below:
- Time: 0.00, %A 20% and %B 80%.
- Time 10.00, %A 60% and %B 40%.
- Time 15.00, %A 90% and %B 10%.
- Time 20.00, %A 20% and %B 80%.
- Use post run time of 10 min.
Note: You must ensure that binary pump pressure reaches initial conditions, and this may differ depending on the void volume of the UHPLC instrument, usually 10 min will be enough. - The typical separation chromatography of the standard is shown in Figure 1 and separation chromatography of the standard in Figure 2.
- You must inject a blank (water only) after every 10-20 injections, depending on the samples. Also you must wash the column every 100-150 samples with 50% methanol in water containing 0.1% formic acid with flow rate of 0.2 ml/min.
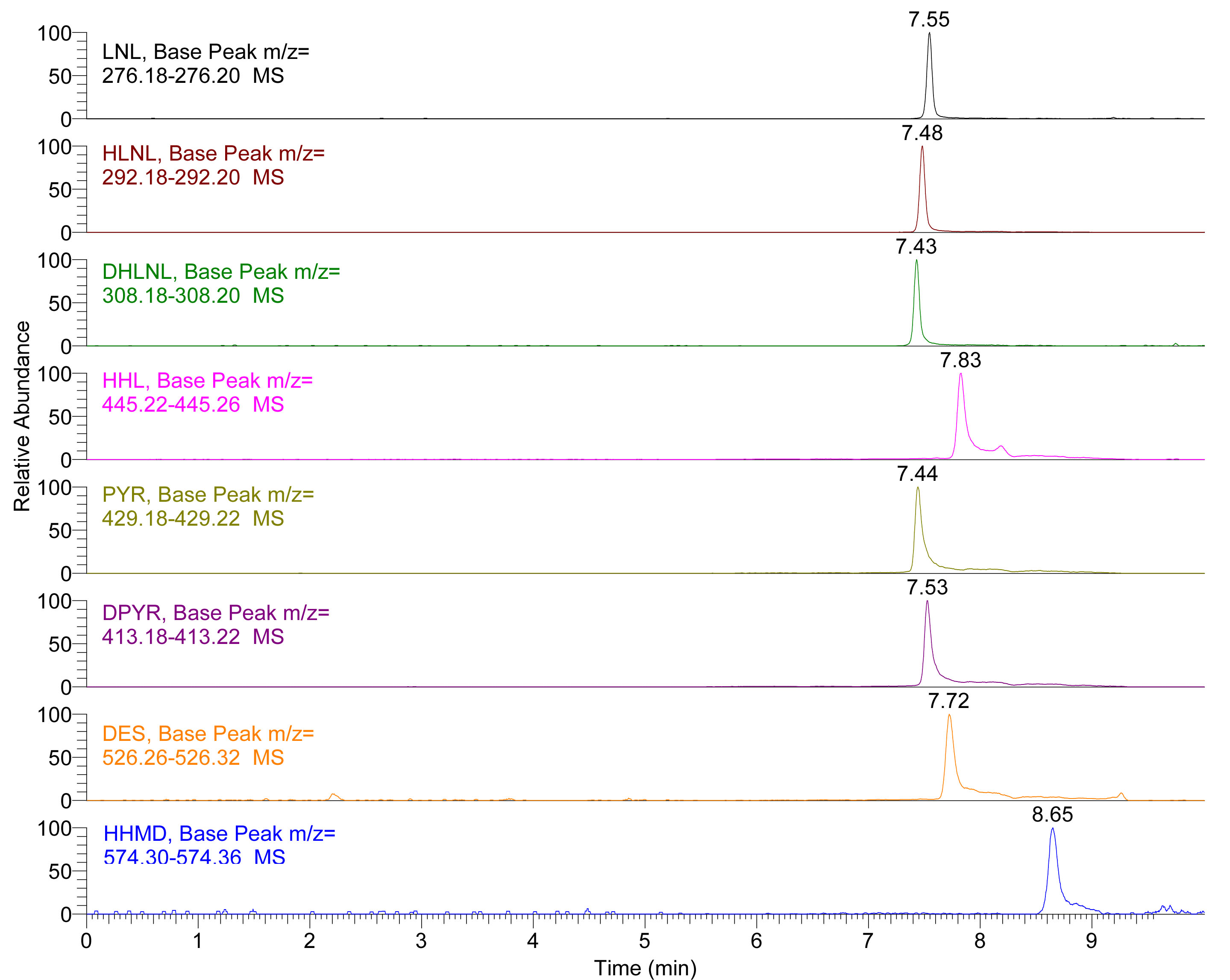
Figure 1. Extracted Ion Chromatogram (EIC) of LNL, HLNL, DHLNL, HHL, PYR, DPYR, DES and HHMD standards. Insets show the extracted mass range of each crosslink.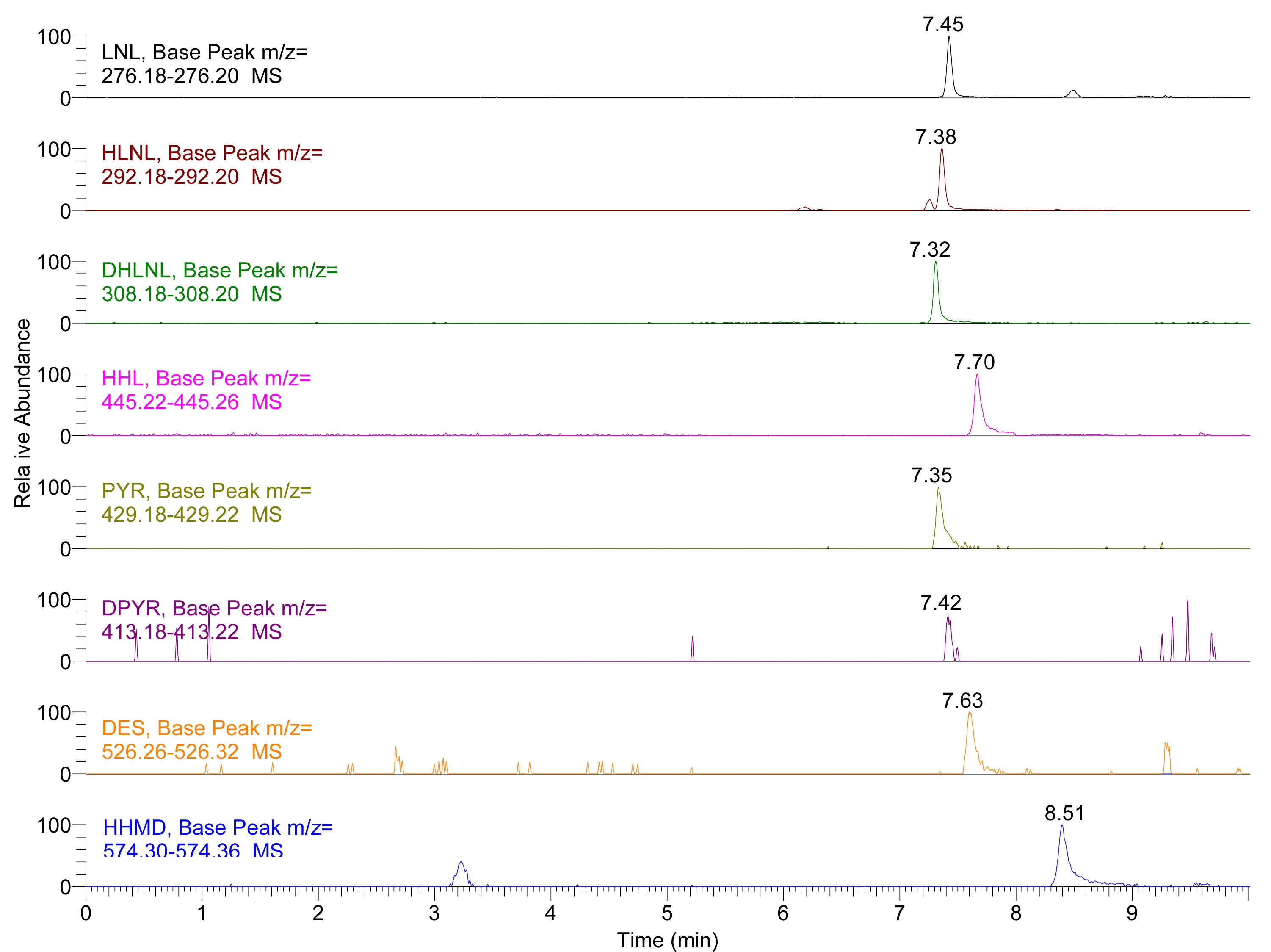
Figure 2. Extracted Ion Chromatogram (EIC) of LNL, HLNL, DHLNL, HHL, PYR, DPYR, DES and HHMD in skin hydrolysates (for the preparation of biological sample hydrolysates see Naffa et al., 2016 and Avery et al., 2009 for more details). Insets show the extracted mass range of each crosslink.
Data analysis
- Open the raw mass spectrometry file on Thermo Xcalibur 4.1.31.9 for qualitative analysis.
- Extract the below monoisotopic masses and observe a ± 0.01 m/z.
- LNL (276.19 m/z).
- HLNL (292.19 m/z).
- DHLNL (308.19 m/z).
- HHL (445.24 m/z).
- PYR (429.20 m/z).
- DPYR (413.20 m/z).
- HHMD (574.33 m/z).
- DES (526.29 m/z).
- There might be small differences in the masses based on the calibration of the mass spectrometry. Always calibrate the mass spectrometry to prevent mass drift (Ask the instrument technician to help you as it requires a specific procedure).
- You can start with a wide mass window (i.e., 292.1000-292.3000 m/z) then refine the window based on the mass measured.
- You can extract the mass of the doubly charged ions for DES and HHMD instead of their singly charged ion mass which will have larger intensity. (Mass of the doubly charged ions is calculated by adding 2.0 to the mass of DES or HHMD then divided by 2, i.e., for HHMD the doubly charged mass is [573.33 + 2.0]/2 = 287.66 m/z).
- Use the peak areas and retention times crosslink quantification (Thermo Xcalibur 4.1.31.9 June 8, 2017 for quantitation analysis using the Genesis algorithm with automated determinations performed on the sample injected).
Note: For analysis of biological samples, identification of crosslinks is performed based on their mass and retention time. This must be always matched with crosslink standard run alongside. Slight variations in the retention times of the crosslinks are expected depending on the sample types, number of runs, washing protocols and age of column.
Recipes
- Solvent A
To make solvent A, measure 1,000 ml of mass spectrometry grade water then add 1.0 ml (1,000 μl) of formic acid - Solvent B
To make solvent B, mix 800 ml of mass spectrometry grade acetonitrile and 200 ml water then add 1.0 ml (1,000 μl) of formic acid
Acknowledgments
This work was supported by NZ Leather and Shoe Research Association (LASRA®), Palmerston North, New Zealand through the Ministry of Business, Innovation and Employment (MBIE) grant number LSRX1701 & 1801.
This protocol was adapted and modified from Naffa et al. (2016 and 2019b).
Competing interests
The authors declare that they have no conflict of interest financial or non-financial.
References
- Arseni, L., Lombardi, A. and Orioli, D. (2018). From structure to phenotype: Impact of collagen alterations on human health. Int J Mol Sci 19(5): e1407.
- Avery, N. C., Sims, T. J. and Bailey, A. J. (2009). Quantitative determination of collagen cross-links. Methods Mol Biol 522: 103-121.
- Gineyts, E., Borel, O., Chapurlat, R. and Garnero, P. (2010). Quantification of immature and mature collagen crosslinks by liquid chromatography-electrospray ionization mass spectrometry in connective tissues. J Chromatogr B Analyt Technol Biomed Life Sci 878(19): 1449-1454.
- Naffa, R., Edwards, P. J. B. and Norris, G. (2019a). Isolation and characterization of collagen type I crosslink from skin: high-resolution NMR reveals diastereomers of hydroxylysinonorleucine crosslink. Amino Acids 51(4): 705-715.
- Naffa, R., Holmes, G., Ahn, M., Harding, D. and Norris, G. (2016). Liquid chromatography-electrospray ionization mass spectrometry for the simultaneous quantitation of collagen and elastin crosslinks. J Chromatogr A 1478: 60-67.
- Naffa, R., Watanabe, S., Zhang, W., Maidment, C., Singh, P., Chamber, P., Matyska, M. T. and Pesek, J. J. (2019b). Rapid analysis of pyridinoline and deoxypyridinoline in biological samples by liquid chromatography with mass spectrometry and a silica hydride column. J Sep Sci 42(8):1482-1488.
- Pinnell, S. R., Krane, S. M., Kenzora, J. E. and Glimcher, M. J. (1972). A heritable disorder of connective tissue. Hydroxylysine-deficient collagen disease. N Engl J Med 286(19): 1013-1020.
Article Information
Copyright
© 2019 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Naffa, R. and Pesek, J. (2019). Separation of Natural Collagen Crosslinks Using Buffer and Ion-pairing Agent Free Solvents on Silica Hydride Column for Mass Spectrometry Detection. Bio-protocol 9(9): e3224. DOI: 10.21769/BioProtoc.3224.
Category
Biochemistry > Protein > Quantification
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











