- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Measuring Protein Synthesis during Cell Cycle by Azidohomoalanine (AHA) Labeling and Flow Cytometric Analysis
Published: Vol 9, Iss 8, Apr 20, 2019 DOI: 10.21769/BioProtoc.3215 Views: 9741
Reviewed by: Gal HaimovichAlexandros C KokotosVarpu Marjomaki

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Analyzing the Properties of Murine Intestinal Mucins by Electrophoresis and Histology
Ran Wang and Sumaira Z. Hasnain
Jul 20, 2017 20576 Views

Analysis of the Ubiquitination and Phosphorylation of Vangl Proteins
Di Feng [...] Bo Gao
Oct 20, 2022 3415 Views

Isoform-specific, Semi-quantitative Determination of Highly Homologous Protein Levels via CRISPR-Cas9-mediated HiBiT Tagging
Kristina Seiler [...] Mario P. Tschan
Jul 20, 2023 2519 Views
Abstract
Protein synthesis is one of the most fundamental biological processes to maintain cellular proteostasis. Azidohomoalaine (AHA) is a non-radioactive and “clickable” amino acid analog of methionine which can be incorporated into newly synthesized proteins. Thus, AHA-labeled nascent proteins can be detected and quantified through fluorescent labeling by "click" chemistry. Here we describe a protocol to measure protein synthesis by AHA labeling and flow cytometry. Taking advantage of gating different cell populations, we provide a typical example of the flow cytometric-based analysis of protein synthesis during the cell cycle. While we used mouse B cells in this protocol this method can be readily applied to any cell types and organisms.
Keywords: Protein synthesisBackground
Traditionally measurement of protein synthesis is performed by pulse labeling of translation products using radiolabeled amino acids such as [35S]methionine and [35S]cysteine but the use of radioactive materials is the major disadvantage. Recent advance in bio-orthogonal chemical reporters such as azides and alkynes allows us to quantitatively monitor and track biomolecules (proteins, lipids or nucleic acids) through click chemistry (Dieterich, 2010). Azidohomoalaine (AHA) (Figure 1) is a methionine analog that contains an azide moiety and is incorporated into newly synthesized proteins. Thus, nascent proteins can be detected through "click" reaction between AHA (azido) and a fluorescent alkyne (Dieterich, 2010).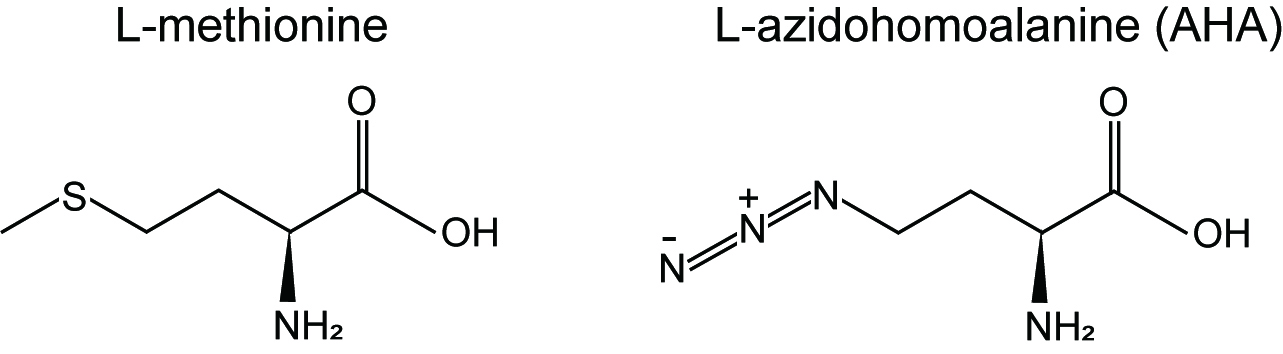
Figure 1. Chemical structures of L-methionine and L-azidohomoalaine (AHA)
Measuring protein synthesis during the cell cycle in mammalian cells has been challenging. To do this, cell synchronization in specific phases of the cell cycle is necessary but the use of drugs such as nocodazole was shown to affect translation (Coldwell et al., 2013; Shuda et al., 2015). To measure protein synthesis without drug treatment, here we used metabolic pulse labeling with AHA in a flow cytometric assay (Kiick et al., 2002; Shuda et al., 2015). In combination with AHA, we used anti-phospho-Histone H3 (Ser10) antibody as a mitotic marker to monitor mitotic translation in mouse B lymphoma cells (19DN) (Sander et al., 2015).
Materials and Reagents
- Pipette tips (Eppendorf, epT.I.P.S.)
- 6-well plate (Thermo Fisher Scientific, BioLite 6-well Multidish, catalog number: 130184)
- 96-well U- or V-bottom cell culture plate (Corning, catalog number: 6902D09 or 6928A17)
- 5 ml round bottom polystyrene FACS tube (Falcon, catalog number: 38007)
- L-Azidohomoalanine (AHA) (Anaspec, catalog number: AS-63669, 500 mM stock solution in deionized water, keep at -20 °C up to at least a few months)
- Methionine-free medium such as Dulbecco’s Modified Eagle’s Medium–high glucose With 4,500 mg/L glucose and sodium bicarbonate, without L-methionine, L-cystine and L-glutamine, liquid, sterile-filtered, suitable for cell culture (Merk, catalog number: D0422)
- Fetal bovine serum, South America origin, dialyzed, sterile filtered (Pan Biotech, catalog number: P30-2102)
- L-Methionine (Merck, catalog number: M9625, make 300 mM stock solution in deionized water, keep at -20 °C up to at least a few months)
- L-Cystine dihydrochloride (Merck, catalog number: C2526, prepare fresh 200 mM (1,000x for DMEM) solution in deionized water)
- GlutaMAX (Thermo Fisher Scientific, catalog number: 35050061, 200 mM stock solution)
- D-PBS (1x), no calcium chloride, no magnesium chloride, sterile-filtered (Thermo Scientific Fisher, catalog number: 14190144)
- Alexa Fluor488 alkyne (Thermo Fisher Scientific, catalog number: A10267)
- Phospho-Histone H3 (Ser10) (D2C8) XP Rabbit mAb (Alexa Fluor 647 Conjugate) (Cell Signaling Technology, catalog number: 3458S)
- (+)-Sodium L-ascorbate (Merck, catalog number: A7631)
- Copper(II)-Sulphate (CuSO4) (Baseclick, catalog number: BCMI-004-50)
- Paraformaldehyde (PFA) (Roth, 335)
- Saponin (Sigma-Aldrich, catalog number: 47036)
- (Optional) Triton X-100 (Sigma-Aldrich, catalog number: T9284)
- Bovine serum albumin (BSA) (Roth, 8076)
- DMSO (Merck, catalog number: D8418)
- EDTA (Merck, catalog number: EDS)
- NaN3 (Merck, catalog number: 71289)
- Cycloheximide (Santa Cruz Biotechnology, catalog number: sc-3508)
- Cell culture medium for AHA labeling (w/o methionine or AHA) (see Recipes)
- Washing solution (see Recipes)
- Fixing solution (see Recipes)
- Permeabilization solution (see Recipes)
- FACS buffer (see Recipes)
Equipment
- (Optional) Multi channel pipette (Eppendorf, catalog number: 3122 000.043 [10-100 μl])
- CO2 Incubator (Thermo Fisher Scientific, catalog number: 51026331)
- Refrigerated centrifuge (Eppendorf, models: 5418 R [fixed angle for tubes], 5804 R [swing out for 96 well plates])
- Flow cytometer (FACS Aria instrument) (BD Biosciences)
Software
- Analysis Software: FlowJo
Procedure
- AHA pulse labeling
- Seed cells in a 6-well plate containing normal cell culture medium so that cells should be 50%-60% confluent next day. Mouse B cells (in suspension) were used in this study but any cell types can be used.
- Next day, starve cells in the methionine-free medium for 30 min.
Note: This step may be omitted to avoid cellular stress. - Start pulse labeling by adding 1 mM AHA or methionine (negative control) to the methionine-free medium and incubate cells for at least 10 min in a 37 °C CO2 Incubator. Cycloheximide treatment (100 μg/ml) can be used as a negative control as well.
- Wash cells with ice-cold PBS. Spin down cells at 500 x g, 4 °C for 3 min and remove the supernatant. Repeat this step once more. For adherent cells such as HEK293, 1 mM EDTA in PBS can be used to harvest cells.
Note: From this step, we used 96-well U- or V-bottom cell culture plate for subsequent steps to increase the throughput. For subsequent steps, use 100-200 μl solution to suspend cells after spinning. - Fix cells in fixing solution (4% PFA in PBS) for 15 min at RT. Spin down cells and remove the supernatant.
- Permeabilize cells with 1% BSA with 0.2% saponin (or 0.25% Triton X-100) in PBS for 15 min. Spin down cells at 500 x g, at RT for 3 min and remove the supernatant.
- Click reaction
- Prepare the click reaction solution (see Recipe 5).
- Incubate samples with 100 μl of the click solution for 30 min at RT. Protect from light.
- Wash cells once with 1% BSA with 0.2% saponin in PBS. Spin down cells and remove the supernatant. In case of Trition X-100 is used for permeabilization, wash cells with 1% BSA in PBS.
- Immunolabeling of a mitotic marker, histone H3 phospho Ser10
- Incubate samples with anti-phospho histone pH3 antibody (1:100, v/v) in 100 μl 1% BSA in PBS for 60 min. Protect from light.
- Wash cells twice with 1% BSA in PBS. Spin down cells and remove the supernatant.
- Resuspend in FACS buffer (1% FBS, 1 mM EDTA, 0.05% NaN3 in PBS) and transfer cells into a 5 ml round bottom polystyrene FACS tube.
- Flow cytometric analysis
- Gate the main cell population by the forward (FSC-A) and side scatter profile (SSC-A) to exclude cellular debris and dead cells (Figure 2A). Gate further the singlet subset by SSC-H and SSC-W (Figure 2B).
- For the subset of cells gated in the Step D1, record AHA-incorporation (y-axis, alexa488) and the mitotic marker (x-axis, alexa647) for at least 10,000 events (Figure 2C). Quantify AHA intensities (y-axis) in interphase and mitotic cells (x-axis) that are separated based on fluorescence intensity of the mitotic marker (see Step D4).
Note: Prepare single color (stained with the Alexa Fluor488-AHA or the Alexa Fluor 647 conjugated antibody) and no color controls for setting FACS parameters and compensation on the flow cytometer. Each compensation control is acquired on the flow cytometer to determine compensation values for each fluorochrome combination. This protocol does not cover detailed instructions for FACS parameter setup. For assistance, please contact your FACS core facility. - Check if the negative control sample (methionine-labeled cells) shows the low level of AHA-incorporation (Figure 2D).
- Quantify the AHA intensities for individual cells (e.g., median intensity) in interphase and mitosis using FlowJo. To do this, open FlowJo and load the files (.fcs) into a workspace. Double-click on a file in the workspace and a plot like Figure 2A will appear. Select subsets for further analyses based on the forward (FSC-A) and side scatter profile (SSC-A) as described in Step D1. Select corresponding populations of interphase and mitosis (see Figure 2C) and click on “Workspace” → “Statistics” → “Median” → “AHA”, which calculates median intensities of AHA signal. Examples of data analysis are shown in Figure 2E.
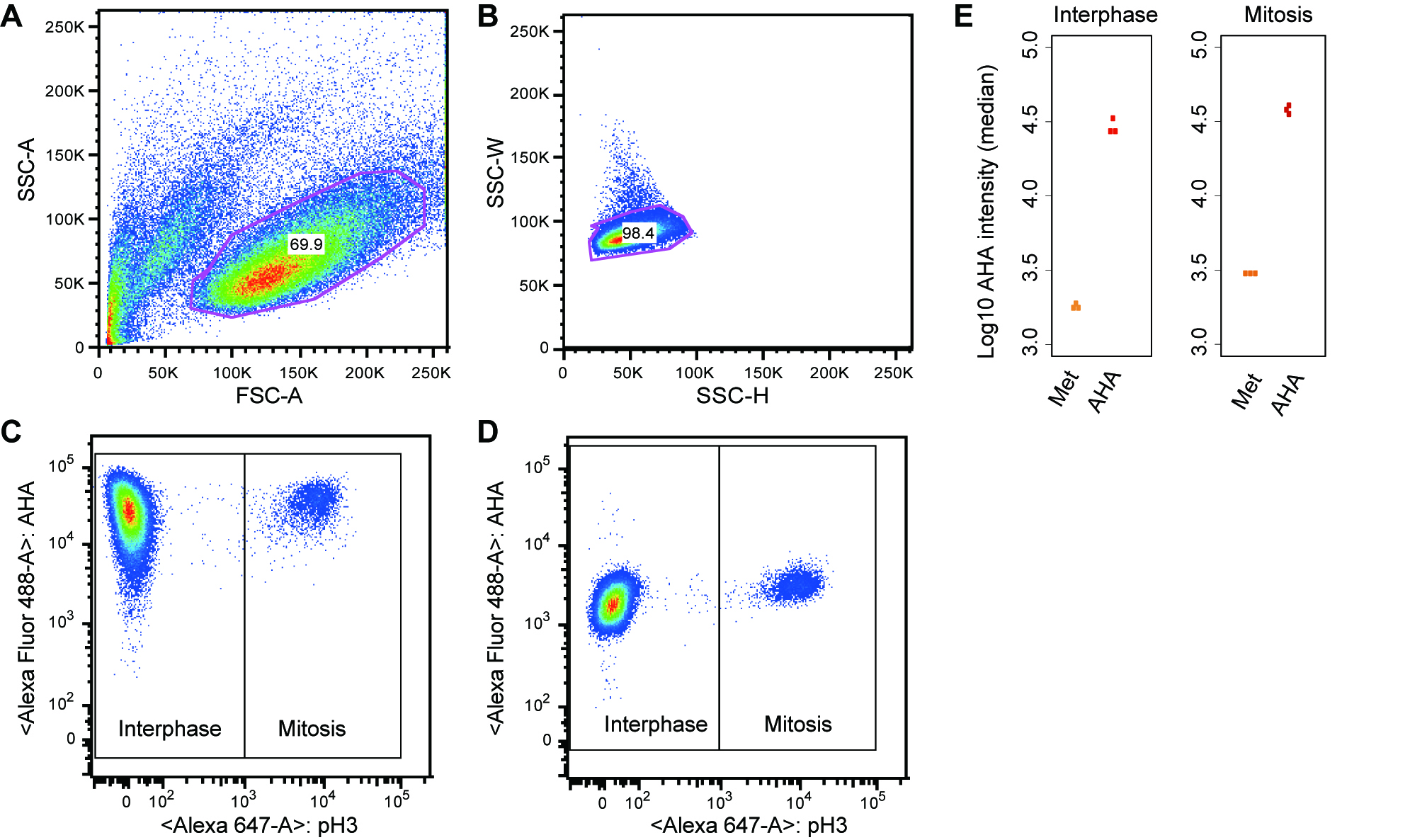
Figure 2. Monitoring global protein synthesis in interphase and mitosis. A and B. Gating strategy. C. The main cell population was further analyzed by dual staining for phospho-H3 S10 and AHA-labeled proteins. Global protein synthesis was monitored by AHA incorporation (y-axis) in interphase and mitosis (based on H3 pS10 staining, x-axis). D. As a negative control, methionine incorporation into proteins instead of AHA was also monitored. E. Examples of quantitative outputs: each dot represents the median AHA intensity calculated from the interphase or mitotic cell population in an independent experiment. The results from three independent experiments are shown.
Recipes
- Cell culture medium for AHA labeling (w/o methionine or AHA)
500 ml of DMEM without methionine, cysteine, glutamine
50 ml of dialyzed serum (normal serum can be used in case use of dialyzed serum affects cell growth.)
Notes:- As long as methionine-free medium is used, the use of normal serum instead of dialyzed serum will not significantly affect pulse labeling results.
- For Cysteine and GlutaMAX, follow the defined concentration of the medium you use.
- Directly add methionine or AHA to the medium just before pulse labeling.
- Washing solution
1% BSA in PBS - Fixing solution
4% PFA in PBS - Permeabilization solution
1% BSA with 0.2% saponin (or 0.25% Triton X-100) in PBS - Click reaction solution
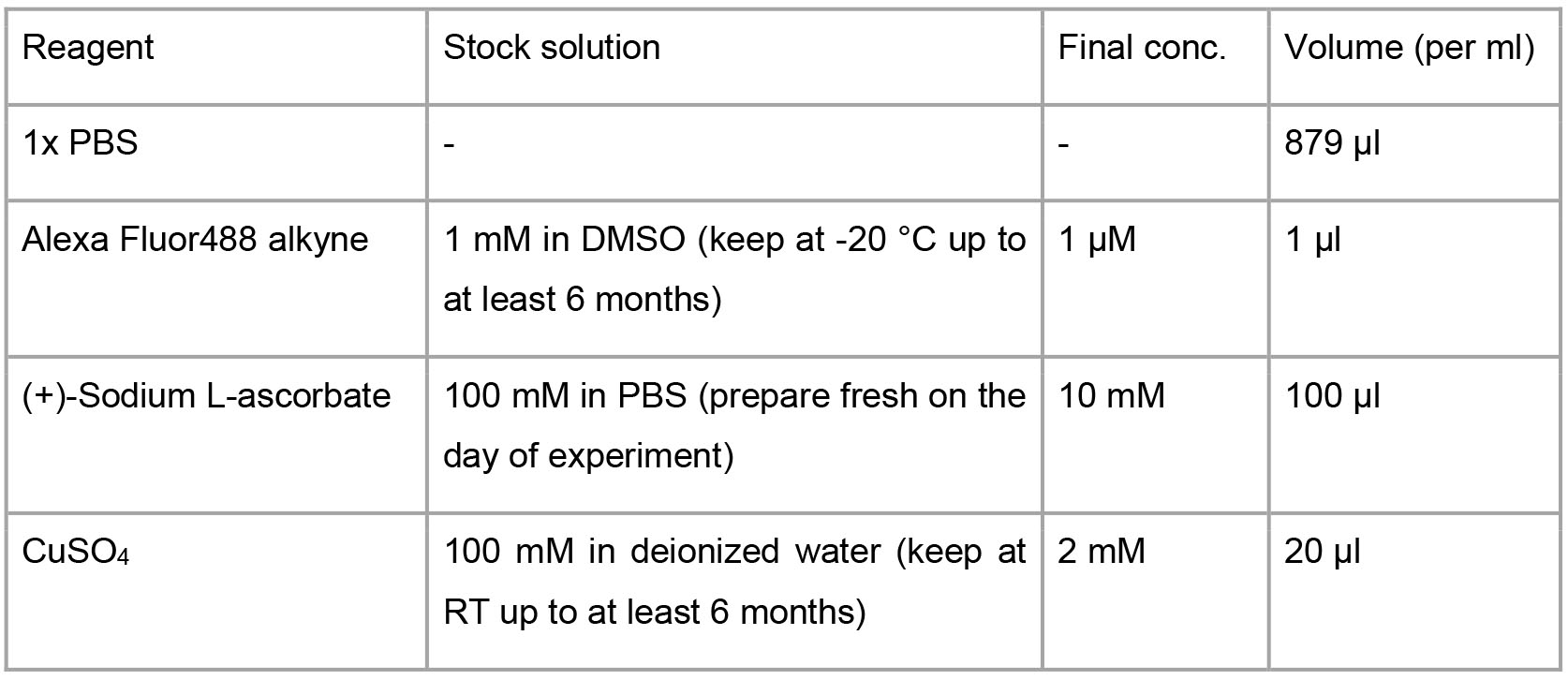
- FACS buffer
1% FBS
1 mM EDTA
0.05% NaN3 in PBS
Notes
You can perform washing, fixing, click reaction and etc. in a 96-well U- or V-bottom cell culture plate to increase the throughput. You can sort the AHA-labeled cells and the sorted cells can be used for many applications including western blot analysis and quantitative analysis using mass spectrometry.
Acknowledgments
We would like to thank Shuda et al. for describing the original protocol (Shuda et al., 2015). We adapted and modified the protocol for this study. We would also like to thank Matthias Selbach (Max Delbruck Center for Molecular Medicine) for critical reading of the manuscript and for his advice.
Competing interests
The authors declare no conflicts of interest or competing interests.
References
- Coldwell, M. J., Cowan, J. L., Vlasak, M., Mead, A., Willett, M., Perry, L. S. and Morley, S. J. (2013). Phosphorylation of eIF4GII and 4E-BP1 in response to nocodazole treatment: a reappraisal of translation initiation during mitosis. Cell Cycle 12(23): 3615-3628.
- Dieterich, D. C. (2010). Chemical reporters for the illumination of protein and cell dynamics. Curr Opin Neurobiol 20(5): 623-630.
- Kiick, K. L., Saxon, E., Tirrell, D. A. and Bertozzi, C. R. (2002). Incorporation of azides into recombinant proteins for chemoselective modification by the Staudinger ligation. Proc Natl Acad Sci U S A 99(1): 19-24.
- Shuda, M., Velasquez, C., Cheng, E., Cordek, D. G., Kwun, H. J., Chang, Y. and Moore, P. S. (2015). CDK1 substitutes for mTOR kinase to activate mitotic cap-dependent protein translation. Proc Natl Acad Sci U S A 112(19): 5875-5882.
- Sander, S., Calado, D.P., Srinivasan, L., Ko¨ chert, K., Zhang, B., Rosolowski, M., Rodig, S.J., Holzmann, K., Stilgenbauer, S., Siebert, R., Bullinger L., Rajewsky K. (2012). Synergy between PI3K signaling and MYC in Burkitt lymphomagenesis. Cancer Cell 22: 167-179.
Article Information
Copyright
© 2019 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Imami, K. and Yasuda, T. (2019). Measuring Protein Synthesis during Cell Cycle by Azidohomoalanine (AHA) Labeling and Flow Cytometric Analysis. Bio-protocol 9(8): e3215. DOI: 10.21769/BioProtoc.3215.
Category
Cancer Biology > General technique > Biochemical assays > Protein analysis
Cell Biology > Cell staining > Protein
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link