- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Tracking Root Interactions System (TRIS) Experiment and Quality Control
Published: Vol 9, Iss 8, Apr 20, 2019 DOI: 10.21769/BioProtoc.3211 Views: 7336
Reviewed by: Yan BaoRaghuram BadmiAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Capturing Z-stacked Confocal Images of Living Bacteria Entering Hydathode Pores of Cauliflower
Aude Cerutti and Alain Jauneau
Oct 20, 2017 9183 Views

Quantification of the Composition Dynamics of a Maize Root-associated Simplified Bacterial Community and Evaluation of Its Biological Control Effect
Ben Niu and Roberto Kolter
Jun 20, 2018 10488 Views

A Quick Method for Screening Biocontrol Efficacy of Bacterial Isolates against Bacterial Wilt Pathogen Ralstonia solanacearum in Tomato
Heena Agarwal [...] Niraj Agarwala
Nov 20, 2020 5546 Views
Abstract
Soil organisms are diverse taxonomically and functionally. This ecosystem experiences highly complex networks of interactions, but may also present functionally independent entities. Plant roots, a metabolically active hotspot in the soil, take an essential part in shaping the rhizosphere. Tracking the dynamics of root-microbe interactions at high spatial resolution is currently limited due to methodological intricacy. In this study, we developed a novel microfluidics-based device enabling direct imaging of root-bacteria interactions in real time.
Keywords: Tracking root interactions system (TRIS)Background
Plant roots play a major role in shaping plant-microbe interactions within the rhizosphere, driving a dynamically changing microenvironment. Recent studies revealed multiple beneficial effects of root-associated bacteria (Hardoim et al., 2008; Weyens et al., 2009; Mei and Flinn, 2010; Santhanam et al., 2015), highlighting the important role of rhizosphere interactions. Tracking these interactions with high spatio-temporal resolution is a challenging task that is primarily limited by available methodologies. Several microfluidic approaches have recently been adapted for the use in the plant sciences (reviewed by Sanati Nezhad, 2014 and Stanley et al., 2016) mainly for root development and physiological studies (Englert et al., 2010; Grossmann et al., 2011; Jiang et al., 2014). The TRIS device presented here builds on these developments to provide a robust platform for microscopy-based studies of the interactions between plant roots and associated microorganisms that can be implemented in a typical experimental biology laboratory (Massalha et al., 2017). The TRIS device enables precise control over environmental parameters within the system while allowing direct observation of dynamic biological processes at high spatial and temporal resolutions. Here we discuss in details the TRIS assembly and operation. Studying root-microbe interactions in this device allows the use of endless combinations of bacterial strains and plant genotypes that could be examined with multiple fluorescent reporters. Altogether, we hope and anticipate that the TRIS experimental system, and other platforms building upon it, will open the way for extensive high-resolution studies of the dynamic root microenvironment and interactions within it.
Materials and Reagents
- Petri dishes (150 mm x 25 mm) (Corning, catalog number: CLS430597)
- Petri dishes (100 mm) (Corning, catalog number: CLS430591)
- 200 µl plastic tips
- Sterile 14 ml polystyrene culture tubes (Corning, catalog number: 352057)
- Glass slides (75 mm x 25 mm) (Thermo, catalog number: 421-004T)
- Adhesive tape (Cellophane tape)
- Millex-HV Syringe Filter Unit, 0.45 µm, PVDF (Merck, Millipore, catalog number: SLHVM33RS)
- 1 ml syringes (BD, catalog number: 309647)
- Polyethylene tubing (OD 1.2 mm) (BD, IntramedicTM, Clay Adams®)
- Regular bamboo toothpicks (can be obtained from any supermarket)
- Parafilm
- 1.5 ml tubes (Eppendorf, catalog number: 0030120086)
- Blunt end needle (McKESSON, catalog number: 8881202363)
- Cotton (can be obtained from any supermarket)
- Biohazard bag (the institute warehouse)
- Arabidopsis seeds (Col-0)
- Bacillus subtilis
- Glycerol stock of Bacillus subtilis NCIB 3610 mCherry reporter strain (obtained from a single colony, stored at -80 °C)
- Polydimethylsiloxane (PDMS) and Curing agent (Sylgard 184 Silicone Elastomer Kit) (Dow Corning Corporation)
- 100% Ethanol (Sigma, catalog number: 34852)
- dH2O (Sigma, catalog number: W4502)
- KOH (Sigma, catalog number: P5958)
- HCl (Sigma, catalog number: H1758) and bleach (Sigma, catalog number: 425044) for seed sterilization.
- Half strength plant growth media, basal salt Murashige and Skoog (MS) (Duchefa Biochemie, catalog number: M0221)
- Plant agar (Duchefa Biochemie, CAS number: 9002-18-0)
- TB buffer (see Recipes)
Equipment
- Forceps
- Scalpel
- 250-ml glass flask
- Pipettes (Eppendorf 20, 200, 1,000 µl)
- Oven
- Hot plate
- OD (600 nm) reader
- 37 °C shaking incubator
- Microfluidic channel mold (see attached CAD file)
- SU-8 2150 (MicroChem Corp.)
- Desiccator with vacuum line
- Harris Unicore hole punch (1 mm) (Ted Pella, catalog number: 15074)
- Hand-held corona generator (Laboratory Corona Treater, Electro-Technic Products, model: BD-20AC)
- Autoclave
- Desiccator
- Inverted optical microscope equipped with dark-field objective (e.g., Olympus)
- Confocal microscope with automated stage system (e.g., Nikon Ti-eclipse)
Software
- Dedicated confocal microscope software
In this study, we used the Nikon confocal and the NIS-element program. - Fiji (ImageJ)
Procedure
- Tracking root interactions system (TRIS) device fabrication and assembly (start a few days before the experiment)
- Microchannel molds (Figure 1A) were prepared by depositing the photoresist SU-8 2150 on silicon wafers using soft lithography techniques (Qin et al., 2010).
- The procedure was applied based on the manufacture guide sheet to achieve 160 µm depth.
Note: Read the manufacturer’s recommendations for proper handling of such viscose material (SU-8 2150). - TRIS device assembly:
- Place the mold into a 150 mm Petri dish with the patterned mold side of the etched photoresist facing up (Figure 1A). Fix the mold to the plate with sticky tape.
- Prepare 25 ml of the PDMS mixture based on the manufacturer’s recommendations. In a plastic weigh-boat, weigh 10 parts silicon elastomer then add 1 part of the curing agent. Mix well the two reagents using a plastic stick.
- Place the dish in a desiccator and degas under vacuum for 1 h. It is important to remove all air bubbles from the PDMS mixture before pouring it to the fixed mold on the dish.
- Blow nitrogen or air to remove any dust that may have accumulated on the mold. Carefully pour the degassed mixture over the silicon master.
- Cure the PDMS for overnight at 60 °C.
Note: Read the manufacturer’s suggestions for curing PDMS. - Using a sharp blade (Scalpel) carefully cut out each adjacent 8/9 features in a single rectangular piece (Figures 1B and 1C). The patterned microchannels will be visible in the PDMS.
- Punch holes at the channel inlets, outlets and the root hole (Figures 1C and 1D) with the 1 mm Harris Unicore hole punch.
- After finishing making the holes, cover both sides of the channels (e.g., with adhesive tape) to protect from dust particles and debris.
- Before binding the PDMS slab to the glass slide, remove the adhesive tape, place the glass slide and PDMS channel side up on a clean flat surface. In order to activate the surfaces of the PDMS and the glass slide, which will lead to covalent bonding of the two materials, we used the hand-held corona generator approximately 1-2 cm above the surface. The surfaces were treated for 30 s uniformly over each surface.
- Carefully place the PDMS device on the treated glass slide with the microchannel pattern facing the glass slide. Apply gentle, uniform pressure by hand to ensure a uniform seal.
- Bake the mounted channel with the glass slide at 100 °C for 20 min directly after sealing.
- Autoclave the mounted microfluidic devices once at 120 °C for 20 min. Store the autoclaved samples sterilely at room temperature conditions.
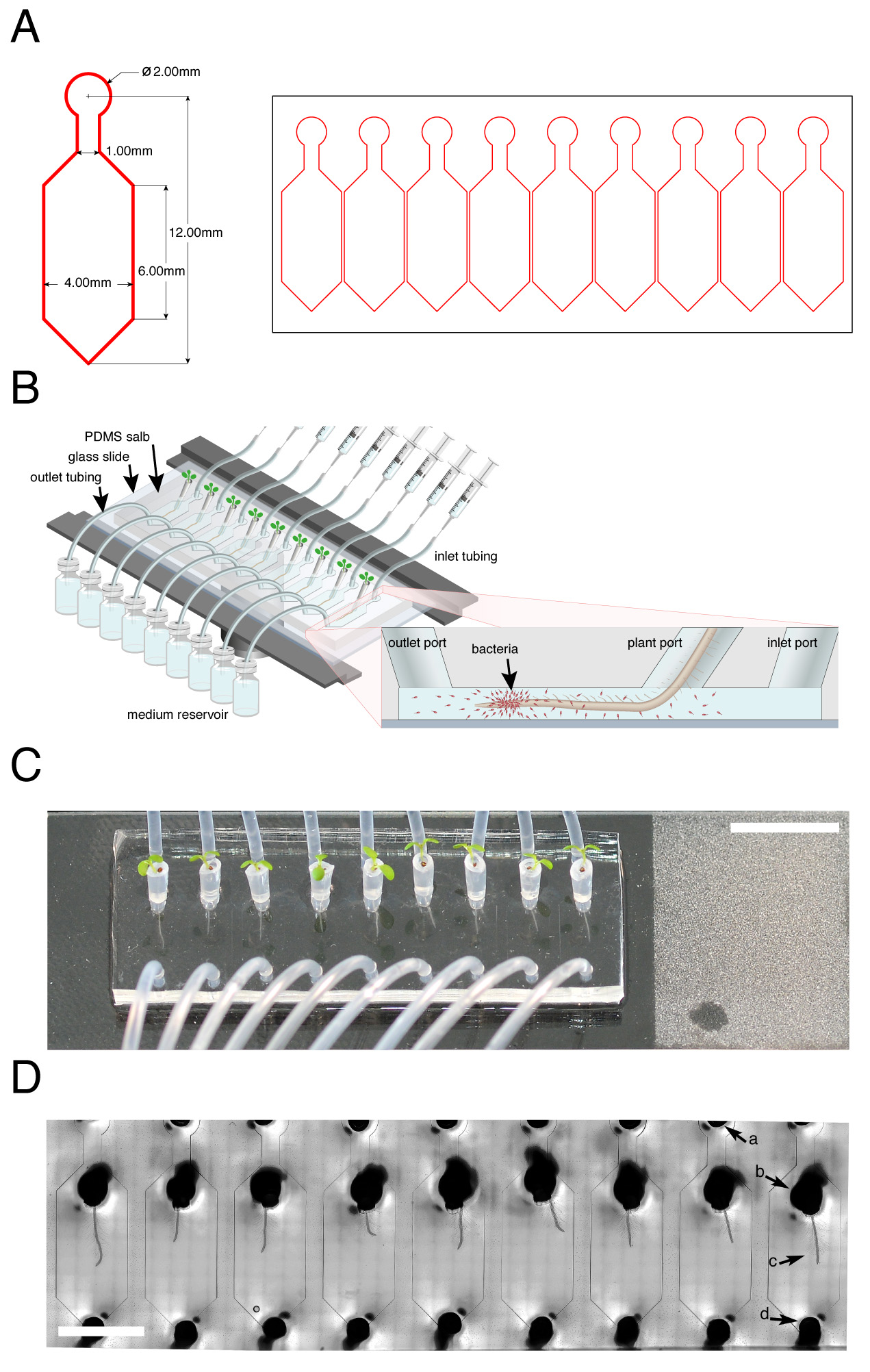
Figure 1. TRIS: a microfluidic device for live tracking of root-bacteria interactions. A. Microfluidic device mask (pattern) design used for the photolithography process to generate an array of nine single microfluidic channels. Left side: Magnification of one single-channel dimensions. B. Illustration of the TRIS device mounted on the microscope stage (dark rim). (Inset) Schematic of a longitudinal section of a microfluidic channel-containing root and bacterial cells (red; not drawn to scale). C. Top view of Arabidopsis seedlings growing in plastic pipette tips attached to the TRIS device. Inlet, plant and outlet ports are attached to the inlet tube, tip with seedling and outlet tube, respectively. Roots are visible as thin white strings extending from the tip ends. (Scale bar: 1 cm). D. Microscopic view of nine Arabidopsis roots growing inside the TRIS microfluidic device captured using bright-field illumination at 10x magnification. Arrows: a, inlet port; b, tip in a root-dedicated port with c, root extending toward the outlet port; d, outlet port. (Scale bar: 5 mm). This figure adapted from Massalha et al. (2017).
- Bacterial culture preparation (start one day before the experiment)
- Place B. subtilis culture in a sterile polystyrene culture tube containing 3 ml TB medium into a shaking incubator at 37 °C, 165 rpm for overnight.
- Four hours before the experiment dilute 1:100 into fresh 3 ml TB medium and incubate again in the same conditions to reach OD600 of 0.6.
- Wash cells twice with MS, and resuspend the cells in MS medium to obtain 106 cells/ml (filter the MS before using to eliminate possible small debris using 0.45 µm PVDF syringe filter).
- Estimate the ratio of swimming cells to static cells by counting the number of the moving cells from a few frames recorded using dark-field microscopy.
- Plant growth (start 5-6 days before the experiment)
- Medium and plastic tip preparation:
- Prepare 0.5x strength basal salt MS. Adjust the pH to 5.8 using KOH. Divide the prepared media into two portions. Add 1% plant agar to one portion.
- Sterilize the two portions of the medium.
- Sterilize 200 µl plastic tips, and bamboo toothpicks, by autoclave.
- After cooling down the medium melt the solidified medium and fill the sterilized 200 µl tips with 5 µl. Allow medium to solidify again. It is important to use the liquid media right before it solidifies at the room temperature (Figure 2A shows plastic tips with solidified medium).
- Before the medium stock solidifies, fill a 90 mm Petri dish with ~30 ml of medium. Allow the medium in the Petri dish to solidify.
- Cut the filled tips with a flamed scalpel just above the level of the solidified medium in the tip in a sterile environment. Use flamed forceps to hold the tips in a sterile 150 mm Petri dish.
- Collect the filled plastic tips with flamed forceps and put them in the solidified agar (in the 90 mm Petri dish prepared in Step C1e). Filled tips fixed in the solidified agar in a Petri dish can be sealed with parafilm and stored at 4 °C for later use.
- In parallel to (1), sterilize Arabidopsis (e.g., Col-0) seeds in chlorine gas for 2 h in a closed desiccator. Place open 1.5 ml tubes with the seeds in a desiccator vessel (in the fume hood). Place a 250-ml glass flask containing 100 ml of bleach into the desiccator and 6 ml of concentrated HCl, immediately close and seal the desiccator with parafilm. After 2 h, open the desiccator in the fume hood and let the chlorine vapor dissipate for several minutes.
- Using a sterile toothpick, transfer one seed aseptically to a plastic pipette tip containing half-strength basal salt MS supplemented with 0.8% plant agar (Figure 1C). Prepare as much tips with seeds as needed for your experiment.
- Stratify seeds by keeping the plates with tips two days in the cold room covered to avoid light.
- Transfer seed-containing tips to a 16-h light/8-h dark period at 23 °C for 3-4 d (or before the growing root penetrates from the end of the plastic tip) (Figure 2A).
- Put the mounted microfluidic devices in a box, in a biohazard bag and sterilize it using autoclave (similar to Step ‘A3l’ in TRIS device assembly section).
- Fill carefully the autoclaved microfluidic device with liquid 0.5x MS to eliminate air bubbles trapped in the system in sterile conditions.
- Place plastic tips with growing seedlings in the autoclaved TRIS device in sterile conditions (Figures 1A and 1B).
- Incubate vertically for an additional 4-5 d before imaging (until the growing root reaches the middle of the TRIS channel). Add liquid medium to the vertical plate to cover the output hole and maintain humidity (Figure 2B). Seal the plate with parafilm to prevent any liquid leakage.
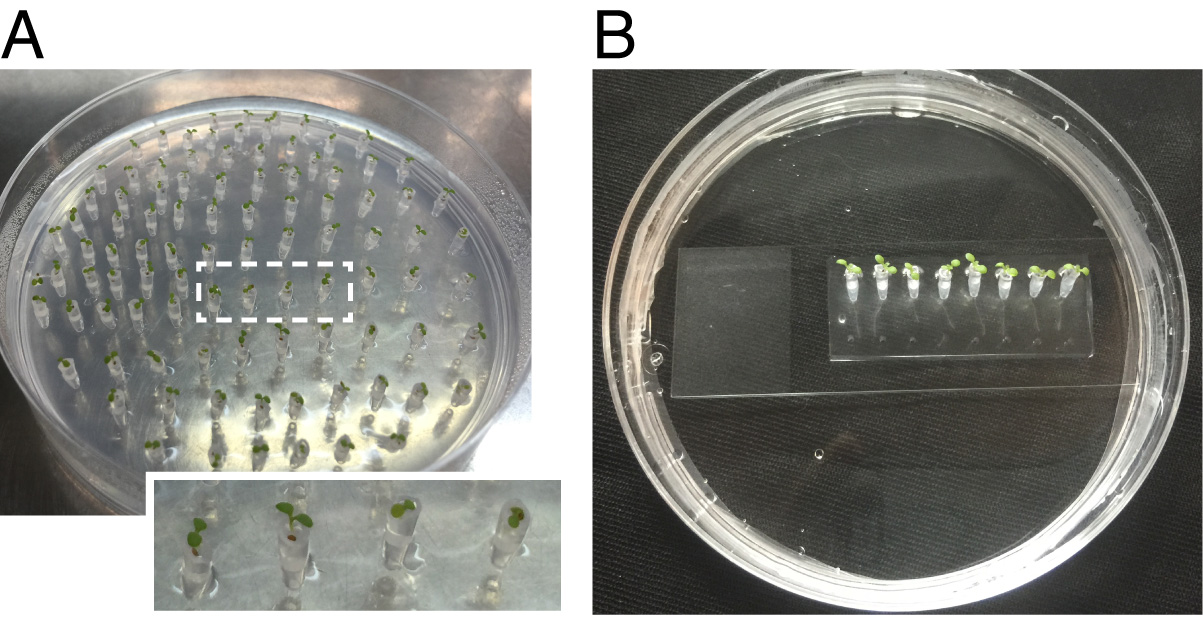
Figure 2. Preparation of Arabidopsis seedlings for tracking root-bacteria interaction using the microfluidics TRIS device. A. Tips of plastic tips filled with solidified agar (the filled agar settled to the same level in all of the tips, indicated by the height of the Arabidopsis seed in the transparent plastic tip) in which Arabidopsis seedlings were germinated placed in agar plates. B. Arabidopsis seedlings in plastic tips fixed to the TRIS device and incubated vertically.
- Medium and plastic tip preparation:
- Mount the TRIS system to the microscope stage (start 2 h before imaging)
- During bacterial incubation time, wash all tubing that will be attached to the microfluidics chambers. Prepare two tubes per channel (long and short tubings, see next steps to determine the length of each tube). Clean all tubing with 70% ethanol to remove possible manufacture contaminants. Wash the tubings twice with dH2O to remove ethanol remaining using a 1 ml syringe.
- Fix a blunt end needle to 1 ml syringe containing washed bacteria in plant medium. Remove air bubbles from the opening of the syringe. Avoid fast fill to prevent air bubbles.
- Connect the long tubing to the blunt end needle and fill the tubes by pushing the piston of the syringe. Make sure the length of the tube is enough to allow flexibility during the assembly of the system on the microscope stage (see Figure 3).
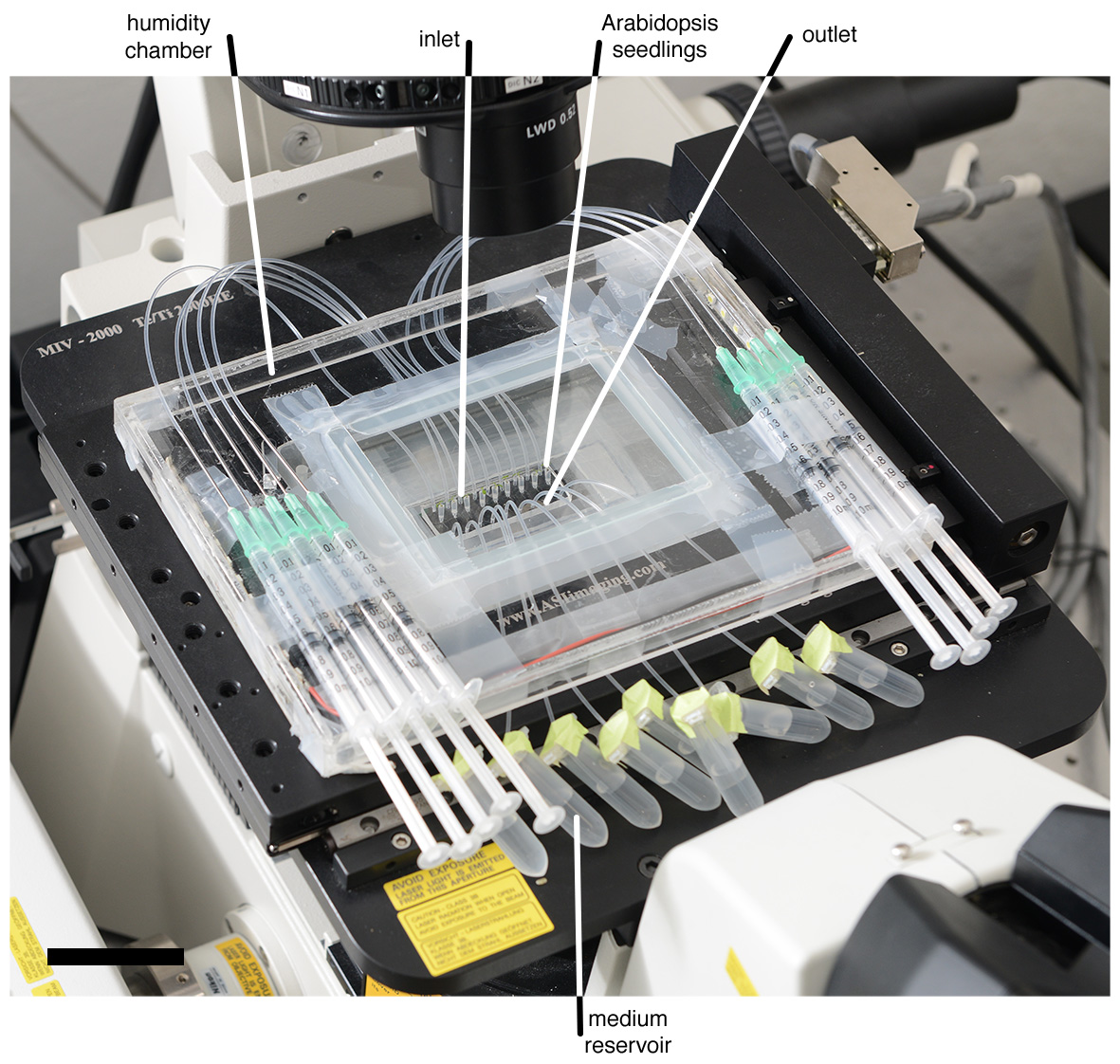
Figure 3. View of the TRIS device mounted onto a microscope stage. The device contains microfluidics channels with Arabidopsis seedlings and the entire setup includes tubing systems, 1 ml syringes, a medium reservoir, and a humidity chamber made of acrylic plastic (Scale bar: 5 cm). Modified from Massalha et al. (2017). - Keep the short tube without filling with liquid.
- Prepare Eppendorf tubes according to the number of microchannels. Make one hole in the lid of the Eppendorf tube using fine forceps and fill with 1 ml MS.
- Quickly, remove the TRIS device from the vertical plate, dry liquid drops with Kimwipe (avoid drying the content of the TRIS device by touching the inlet/outlet holes), fix the TRIS device to the microscope stage holder (preferable slide holder without openings to keep humid conditions).
- Insert the long tube connected to the syringe to the inlet hole and the short tube to the output hole of each microchannel. Apply enough pressure to force the tubing roughly half-way from the PDMS surface to the glass slide but carefully to avoid cracks in the PDMS inlet and outlets.
- Quickly cover the device with a transparent cover to keep humidity (Figure 3).
- Put inside the transparent cover Petri dish a wet cotton to keep the humidity.
- Dip the second end of the output tube (short tube) into an Eppendorf tube filled with 1 ml MS media.
- Set the microscope parameters:
- Scanning (x, y)-coordinates of each channel (stitching parameters should be tested in a preliminary experiment).
- Exposure time of all used channels (should be tested in a preliminary experiment).
- Number of cycles.
- Cycle intervals.
- Find the mid-section of the root. Set the ± 40 µm z-stacks.
- Introduce the bacteria to the microfluidic channel by pushing the piston of the syringe slowly. Continue pushing the piston until the outlet tube will be filled.
- Immediately after finishing Step D9, start scanning that microchannel.
- After Step D10 is finished, pause the microscope, introduce the bacteria to the next microchannel, and then continue scanning.
- Keep doing Steps D12 to D14 until the last microchannel.
- Image analysis
- Fiji is used to visualize the results and generate the summarizing movies we used the LOCI package to open bio-microscopy formats) (Schindelin et al., 2012).
- Extended depth of field package (Fiji, http://imagej.net/Extended_Depth_of_Field) or FStack function from the File Exchange repository (MathWorks) is used to generate a single EDF image from the different z-stacks.
- Matlab is used to segment the EDF images based on the brightness of the bright field channel in each image and then to calculate the relative intensity of the bacterial signal.
Data analysis
The main outcome of the TRIS device is the bacterial density as functions of location and time. The relative intensity of the bacterial fluorescence is used to estimate the density of the bacteria and calculated by subtracting the background for each time point from the region of interest. Other parameters can be calculated using the TRIS device, i.e., root elongation and bacterial dynamics. Data analysis at this stage become highly depended on the biological question.
Notes
The captured volume of the TRIS device is only ~6.4 μl per plant, thus makes plants sensitive to environmental permutations. It is highly recommended to track the growth conditions, especially when moving the plants to the TRIS device.
Recipes
- TB buffer
1% (w/v) tryptone
0.5% (w/v) NaCl
Autoclave
Acknowledgments
This protocol is adapted from Massalha et al. (2017). This research was supported as part of a PhD funded by a Planning & Budgeting Committee of the Council of Higher Education of Israel personal grant (to H.M.).
References
- Englert, D. L., Manson, M. D. and Jayaraman, A. (2010). A microfluidic device for quantifying bacterial chemotaxis in stable concentration gradients. J Vis Exp(38) pii: 1779.
- Grossmann, G., Guo, W. J., Ehrhardt, D. W., Frommer, W. B., Sit, R. V., Quake, S. R. and Meier, M. (2011). The RootChip: an integrated microfluidic chip for plant science. Plant Cell 23(12): 4234-4240.
- Hardoim, P. R., van Overbeek, L. S. and Elsas, J. D. (2008). Properties of bacterial endophytes and their proposed role in plant growth. Trends Microbiol 16(10): 463-471.
- Jiang, H., Xu, Z., Aluru, M. R. and Dong, L. (2014). Plant chip for high-throughput phenotyping of Arabidopsis. Lab Chip 14(7): 1281-1293.
- Massalha, H., Korenblum, E., Malitsky, S., Shapiro, O. H. and Aharoni, A. (2017). Live imaging of root-bacteria interactions in a microfluidics setup. Proc Natl Acad Sci U S A 114(17): 4549-4554.
- Mei, C. and Flinn, B. S. (2010). The use of beneficial microbial endophytes for plant biomass and stress tolerance improvement. Recent Pat Biotechnol 4(1): 81-95.
- Qin, D., Xia, Y. and Whitesides, G. M. (2010). Soft lithography for micro- and nanoscale patterning. Nat Protoc 5(3): 491-502.
- Sanati Nezhad, A. (2014). Microfluidic platforms for plant cells studies. Lab Chip 14(17): 3262-3274.
- Santhanam, R., Luu, V. T., Weinhold, A., Goldberg, J., Oh, Y. and Baldwin, I. T. (2015). Native root-associated bacteria rescue a plant from a sudden-wilt disease that emerged during continuous cropping. Proc Natl Acad Sci U S A 112(36): E5013-5020.
- Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., Preibisch, S., Rueden, C., Saalfeld, S., Schmid, B., Tinevez, J. Y., White, D. J., Hartenstein, V., Eliceiri, K., Tomancak, P. and Cardona, A. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods 9(7): 676-682.
- Stanley, C. E., Grossmann, G., i Solvas, X. C. and deMello, A. J. (2016). Soil-on-a-Chip: microfluidic platforms for environmental organismal studies. Lab Chip 16(2): 228-241.
- Weyens, N., van der Lelie, D., Taghavi, S. and Vangronsveld, J. (2009). Phytoremediation: plant-endophyte partnerships take the challenge. Curr Opin Biotechnol 20(2): 248-254.
Article Information
Copyright
© 2019 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Massalha, H., Korenblum, E., Shapiro, O. H. and Asaph, A. (2019). Tracking Root Interactions System (TRIS) Experiment and Quality Control. Bio-protocol 9(8): e3211. DOI: 10.21769/BioProtoc.3211.
Category
Plant Science > Plant physiology > Biotic stress
Microbiology > Microbe-host interactions > In vivo model > Plant
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









