- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
A Radioisotope-free Oligosaccharyltransferase Assay Method
Published: Vol 9, Iss 5, Mar 5, 2019 DOI: 10.21769/BioProtoc.3186 Views: 6337
Reviewed by: Alessandro DidonnaAswad KhadilkarRoger Salvacion Tan

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
![[2-<sup>3</sup>H]Mannose-labeling and Analysis of N-linked Oligosaccharides](https://en-cdn.bio-protocol.org/imageup/arcimg/20170718074630401.jpg?t=1770651681)
[2-3H]Mannose-labeling and Analysis of N-linked Oligosaccharides
Marina Shenkman [...] Gerardo Z. Lederkremer
Jul 20, 2017 7824 Views

A Robust, One-step FRET Assay for Human Heparanase
Jyothi C. Sistla and Umesh R. Desai
Sep 5, 2019 4660 Views

Analysis of Lipid-linked Oligosaccharides Synthesized in vivo in Schizosaccharomyces pombe
Ayelen Valko [...] Cecilia D’Alessio
Sep 20, 2022 2228 Views
Abstract
Glycosylation of asparagine residues is widespread in Eukarya, and occurs in virtually all Archaea and some eubacterial species. A membrane-bound enzyme, oligosaccharyltransferase, catalyzes the transfer of an oligosaccharide chain from a sugar donor (lipid-linked oligosaccharide, LLO) to an asparagine residue in the consensus sequence, Asn-X-Ser/Thr (X ≠ Pro), in proteins. The in vitro oligosaccharyl transfer assay reaction mixture contains a detergent-solubilized oligosaccharyltransferase (OST), a sugar donor LLO, and a sugar acceptor peptide. Previous assay methods are problematic, in terms of the use of radioactive compounds and the cumbersome separation procedures using lectin binding or two-phase partitioning. Here, we describe a new oligosaccharyl transfer assay method, which is radioisotope-free and relies on a different separation mechanism. The glycopeptide products are separated from unreacted peptides by SDS-PAGE. A fluorescent dye is attached to the peptide substrate during custom peptide synthesis. The fluorescent imaging of the SDS-PAGE gels ensures high sensitivity and quantitative performance. The user-friendly PAGE format is particularly suitable for presentation in scientific papers. For illustrative applications, time-course and peptide library experiments are shown.
Keywords: OligosaccharyltransferaseBackground
Glycosylation of asparagine residues, which is also referred to as N-glycosylation, is one of the most ubiquitous protein modifications (Cherepanova et al., 2016). N-glycosylation occurs not only in Eukarya, but also in most Archaea (Larkin and Imperiali, 2011). N-glycosylation is also found in some species in Eubacteria. In the entire complex N-glycosylation process, the transfer of an oligosaccharide chain from a sugar donor to asparagine residues in proteins is the defining event (Figure 1). This reaction is catalyzed by a membrane-bound enzyme, oligosaccharyltransferase (OST or OTase). The acceptor asparagine residues, in principle, reside in the consensus sequence Asn-X-Thr or Asn-X-Ser (or Asn-X-Cys in rare cases), where X can be any amino acid residue except Pro. Eubacterial OST uses an extended version of the 5-residue sequon, Asp-X-Asn-X-Ser/Thr or Glu-X-Asn-X-Ser/Thr (X ≠ Pro). Asp is preferable to Glu at the -2 position, even though the acidic residue at the -2 position is not absolutely required in some eubacterial species (Schwarz et al., 2011). The sugar donor is a type of glycolipid, referred to as a lipid-linked oligosaccharide (LLO). The chemical structure of LLO is an oligosaccharide chain attached to a lipid-phospho carrier. The lipid part is dolichol in Eukarya and Archaea, and undecaprenol in Eubacteria. The phosphate group is basically diphosphate, but some archaeal species belonging to Euryarchaeota (an ancient phylum of Archaea) use monophosphate (Taguchi et al., 2016).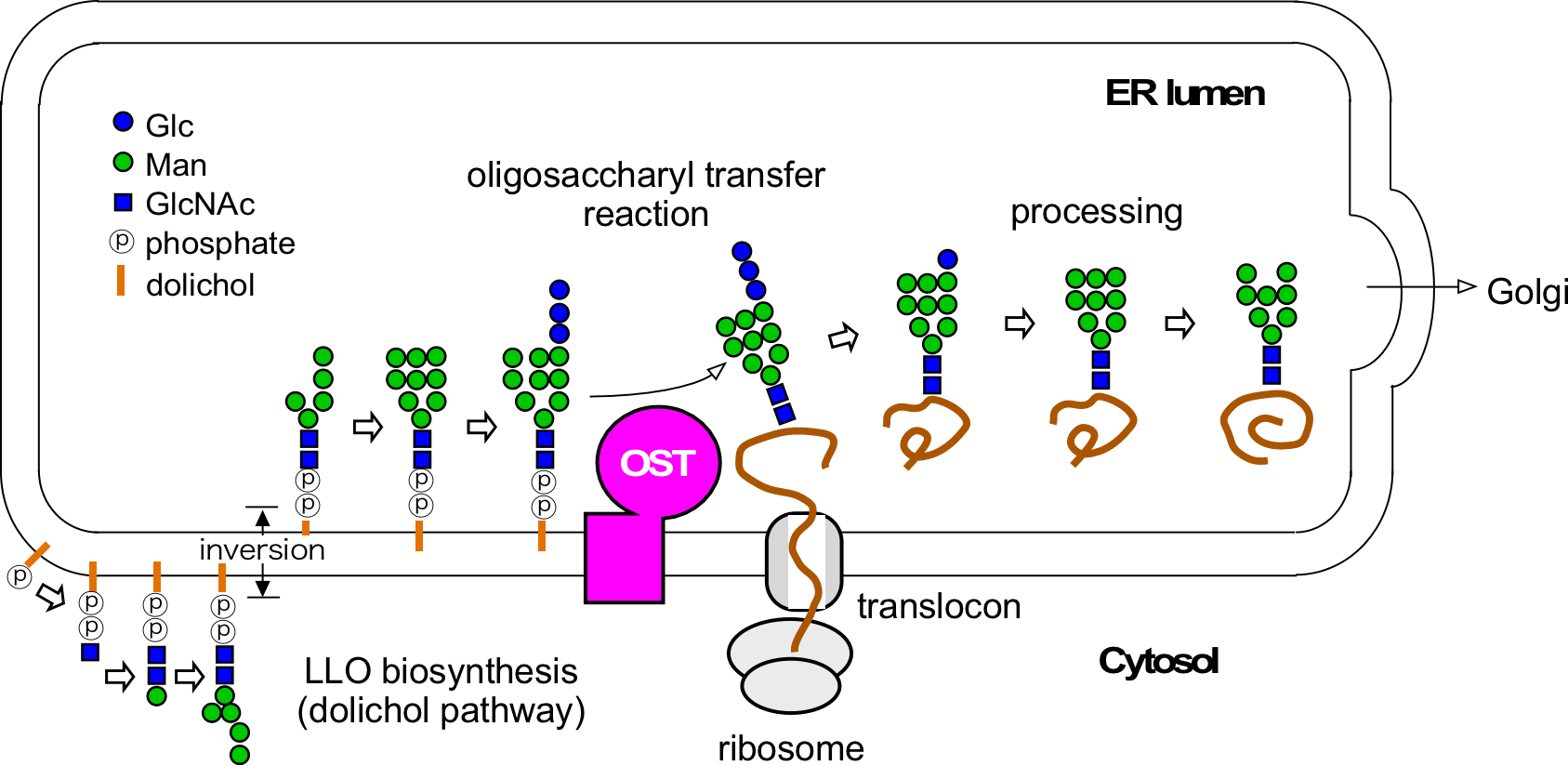
Figure 1. Overview of the biosynthesis of N-glycans attached to proteins. The oligosaccharyltransferase catalyzes the transfer of the oligosaccharide chain from LLO to asparagine residues in nascent proteins on the luminal side of the eukaryotic ER. OST: oligosaccharyltransferase. LLO: lipid-linked oligosaccharide.
The in vivo oligosaccharyl transfer activity is measurable by analyzing the extent of oligosaccharide modification of a specified protein in cells. The amino-acid variants of model glycoproteins can be used to analyze the efficiency of the oligosaccharyl transfer reaction around the N-glycosylation site. However, it is difficult to analyze the effects of amino-acid variation on native oligosaccharyl transfer activity in cells, because the N-glycosylation is essential in eukaryotic cells and rather vital in prokaryotic cells. In general, in-cell studies are difficult to set up. In an exceptional case, the whole N-glycosylation system (the LLO biosynthetic pathway, the OST enzyme, and the substrate protein) of the eubacterium Campylobacter was transferred to Escherichia coli cells (Wacker et al., 2002).
The reaction solution for in vitro oligosaccharyl transfer assay is a mixture of a detergent-solubilized OST enzyme, a sugar donor LLO, and a sugar acceptor peptide. Chromatographically purified LLO is preferred, but crude fractions after two-phase partitioning with a chloroform/methanol/water system are usable. In principle, OST strictly recognizes the chemical structure of LLO. As for eukaryotic OSTs, the chemical structure of LLO (Glc3Man9GlcNAc2-PP-dolichol) is common, and thus the human OST enzyme can utilize LLO isolated from yeast cells. In contrast, the archaeal LLOs are quite structurally diverse and must be isolated from the same or closely related species for the oligosaccharyl transfer assay.
One previous assay method is based on the lectin binding of radioactive glycopeptide products after the oligosaccharyl transfer reaction with a 125I-labeled peptide substrate (Geetha-Habib et al., 1990). Another assay method is based on the recovery of radioactive glycopeptide products in the upper water phase, in two-phase partitioning after the oligosaccharyl transfer reaction with 14C-labeled LLO (Bause and Hettkamp, 1979; Sharma et al., 1981). The two methods are problematic, in terms of the use of radioactive compounds and the cumbersome separation procedures.
Therefore, we have developed a new oligosaccharyl transfer assay method, which is radioisotope-free and highly efficient, by using PAGE as the separation mechanism (Figure 2) (Kohda et al., 2007). The bulky oligosaccharide chain in glycopeptides decreases the rate of migration during electrophoresis, due to the sieving action of the polyacrylamide gel. The glycopeptide products are detected and quantified by fluorescent imaging of the PAGE (polyacrylamide gel electrophoresis) gels. A fluorescent dye for detection is attached to a peptide during peptide synthesis. The minimal detectable quantity of the glycopeptide is 1 fmol in routine assay conditions. The excellent linearity over three orders of magnitude for fluorescently labeled peptide amounts up to 1,000 fmol enabled us to quantify the glycopeptide product accurately, for the determination of the Km and Vmax values for peptide substrates (Igura and Kohda, 2011a). At 12 years after its development, the radioisotope-free oligosaccharyltransferase assay method is now widely used in many laboratories.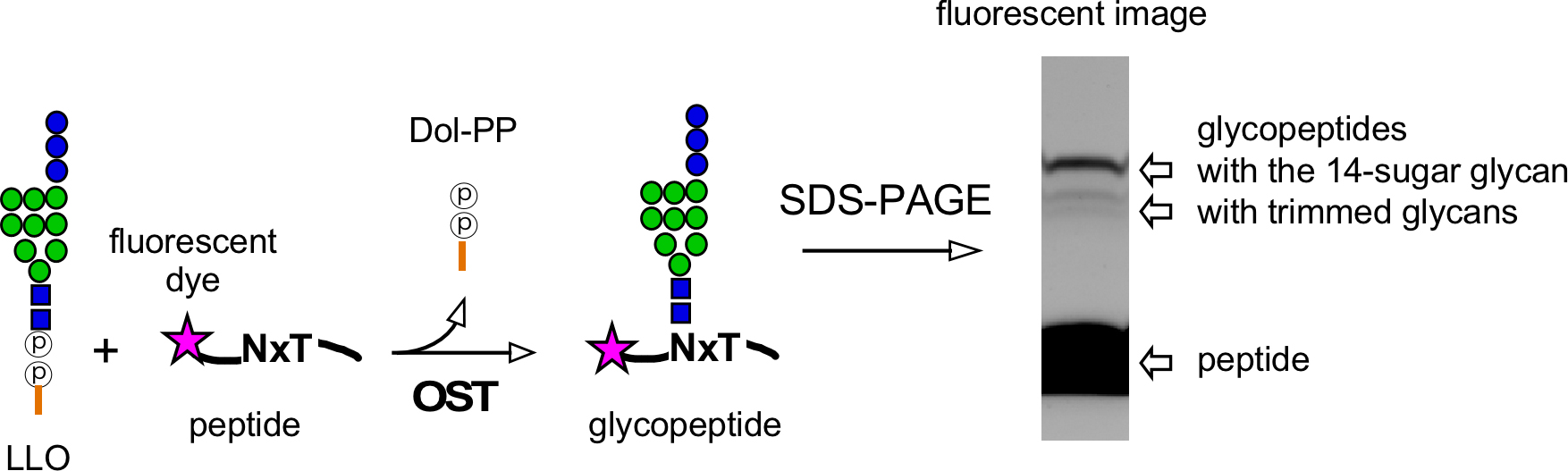
Figure 2. Radioisotope-free assay for the oligosaccharyltransferase activity. Oligosaccharyl transfer assay method using a fluorescently labeled peptide for detection and a separation mechanism based on PAGE (polyacrylamide gel electrophoresis). OST: oligosaccharyltransferase. LLO: lipid-linked oligosaccharide. Dol-PP: dolichol-pyrophosphate
Materials and Reagents
- Labwares
- Pipette tips, 10 μl and 200 μl (Gilson, Diamond tips D10 and D200, catalog numbers: F161630 and F161930)
- Conical polypropylene centrifuge tubes, 15 ml and 50 ml (Thermo Fisher, Nunc, catalog numbers: 339650 and 339652)
- 1.5 ml polypropylene tubes (Eppendorf, 3810X, catalog number: 0030125150)
- Gloves (Ansell, Microflex, catalog number: NPG-INT)
- OST protein and two substrates
- Oligosaccharyltransferase enzyme, isolated from natural sources or recombinantly expressed, purified in the presence of detergent (see Notes for preparation)
- Sugar donor LLO, (partially) purified from cultured cells (see Notes for preparation)
- Sugar acceptor peptide containing the N-glycosylation sequon in the amino acid sequence and a fluorescent dye attached to the N- or C-terminus for detection (see Notes for design and preparation)
- Reagents, solutions, and buffers
- Precast PAGE gels, 15%-25% gradient gel (size 85 x 90 x 0.9 mm3) (Cosmo Bio, MULITGEL II Mini 15/25 (17W), catalog number: DCB-414923)
- Trizma base [tris(hydroxymethyl)aminomethane] (Sigma-Aldrich, catalog number: T1503)
- HCl (Sigma-Aldrich, catalog number: H1758)
- MgCl2 hexahydrate (Sigma-Aldrich, catalog number: M2670)
- MnCl2 tetrahydrate (Sigma-Aldrich, catalog number: 221279)
- Dithiothreitol (DTT) (Sigma-Aldrich, catalog number: 43819)
- Tween 20 (Sigma-Aldrich, catalog number: P1379)
- Lyophilized peptide powder (custom order, TORAY)
- Triton X-100 (Sigma-Aldrich, catalog number: X100)
- N-dodecyl-β-D-maltoside (DDM) (DOJINDO, catalog number: D316)
- Chloroform (Sigma-Aldrich, catalog number: C2432)
- Methanol (Sigma-Aldrich, catalog number: 34860)
- SDS (Sigma-Aldrich, catalog number: L3771)
- Glycerol (Sigma-Aldrich, catalog number: G5516)
- Bromophenol blue (Sigma-Aldrich, catalog number: B0126)
- Glycine (Sigma-Aldrich, catalog number: G8898)
- (Optional) EDTA (ethylenediaminetetraacetic acid) (Sigma-Aldrich, catalog number: EDS)
- 10:10:3 (v/v/v) chloroform:methanol:water (see Recipes)
- 5x SDS-PAGE sample loading buffer (see Recipes)
- Running electrophoresis buffer (see Recipes or Sigma-Aldrich, 10x concentrate, catalog number: T7777-1L)
Equipment
- Micropipettes (Gilson, Pipetman P2, P20, P200, catalog numbers: F144801, F123600, F123601)
- Electronic pipettes (Gilson, Pipetman P10M, P100M, catalog numbers: F81022, F81013)
- 4 °C refrigerator (PHC, MPR-1014)
- Refrigerated laboratory tabletop centrifuge (Eppendorf, Centrifuge 5418R, catalog number: 5401000030)
- Water-bath type sonicator (Branson, Bransonic 2800 ultrasonic cleaner)
- Aluminum heating/cooling block for 1.5 ml tubes (Sigma-Aldrich, catalog number: Z740272-1EA)
- Incubator, natural convection oven or forced air flow convection oven (YAMATO, IC402, catalog number: 211195 or EYELA, NDO-420 or WFO-420, catalog numbers: 252240 or 252280)
Note: Alternatively, a water bath-type incubator and PCR equipment can be used. - Vacuum concentrator that withstands organic solvent (Thermo Fisher, Savant SpeedVac SPD120P1, catalog number: SPD120P1-115/230)
- Cassette electrophoresis unit for PAGE (Cosmo Bio, Model DPE-1020 for Mini Gel, DCB-303111)
- Power supply for PAGE (Bio-Rad, PowerPac Basic, catalog number: 1645050)
- LAS-3000 multicolor gel image analyzer (Fuji Film)
- LED (Green 520 nm EPI) illuminator (for TAMRA dye) and LED (Blue 460 nm EPI) illuminator (for 5-CF/5-FAM dye)
- 575DF20 filter (for TAMRA) and Y515-Di filter (for 5-CF/5-FAM)
- Iris 0.85
- Air (gas) duster (Orientech, catalog number: AD400FL)
- Fume hood (Yamato Scientific, catalog number: RFS-120SZ)
- -20 °C freezer (PHC, catalog number: MDF-MU500H-PJ)
Software
- ImageGauge software (Fuji Film)
Note: Alternatively, any software installed in other gel imagers can be used.
Procedure
- Oligosaccharyl transfer reaction
- Preparation of reaction buffer solution
Combine the following reagents in a 1.5 ml polypropylene tube:
10 μl of 1 M Tris-HCl, pH 7.5
20 μl of 100 mM MnCl2
2 μl of 100 mM DTT
168 μl of 0.1% (v/v) Triton X-100 or 0.1% (w/v) DDM (n-dodecyl-β-D-maltoside)
The total volume is 200 μl
Note: In the original protocol, 0.02% (v/v) Tween 20 was used (Kohda et al., 2007). We now routinely use Triton X-100 or DDM for better LLO solubilization. MgCl2 may be replaced by MnCl2. The solution becomes slightly brown upon the addition of DTT, due to the formation of manganese dioxide, but it poses no problem for the assay. The omission of DTT has an insignificant effect on the assay, but the inclusion of DTT may be expected to clarify the migration image by its denaturing effects on viscous contaminants in the reaction mixture solution. - Solubilization of LLO
- The volume of the LLO stock in 10:10:3 (v/v/v) chloroform:methanol:water is calculated by 10 μl (corresponding to 0.1-10 pmol of LLO) x "the number of samples plus 1 extra for the plastic tubes". The recommended amount of LLO is 10 pmol per 10 μl for quantitative experiments. Usual number of samples is in the range from 10 to 20, in consideration of the number of lanes of one PAGE gel.
- Transfer the LLO solution to a 1.5 ml polypropylene tube and dry it in a SpeedVac concentrator. Temperature is set between 30 °C and 37 °C.
- Add the reaction buffer solution to the dried LLO in the tube. The volume of the reaction buffer solution is calculated by 8 μl x "the number of samples plus 1 extra for the plastic tubes".
- Sonicate the solution in a bath-type sonicator in continuous mode for 5 min at room temperature, and vortex vigorously to dissolve the LLO.
- Centrifuge at 16,000 x g, 5 min, at 4 °C to remove insoluble materials.
- Transfer the supernatant to a new tube, and add the detergent-solubilized OST to the supernatant. The volume of the detergent-solubilized OST is calculated by 1 μl (corresponding to 0.01-1 pmol of the enzyme) x "the number of samples plus 1 extra for the plastic tubes".
- Nine μl aliquots of the supernatant are pipetted into 1.5 ml polypropylene tubes, precooled on ice. The use of an electronic pipette is highly recommended for accurate dispensing.
- Starting and stopping the reaction
- Add 3 μl (corresponding to 30 pmol) of 10 μM TAMRA-acceptor peptide solution to each polypropylene tube. This allows time-course analyses and peptide library experiments to be performed. Centrifuge briefly to spin down the content, tap the tube with a finger a few times for mixing, and centrifuge again.
- Place the tubes on ice. In the case of a large number of tubes, it is convenient to use an aluminum heating/cooling block for 1.5 ml tubes.
- Transfer the tubes (in the aluminum block) to an incubator. Incubate at the desired temperature (30 °C for the yeast and Campylobacter OSTs, 65 °C for thermostable OSTs from thermophilic archaea). The incubation time should be chosen within a time frame corresponding to the linear part of the progress curve, to measure the initial rate of the reaction (Figure 3). Typical incubation time is 30 min to 2 h. At a high temperature, it is important to heat the tubes uniformly to prevent water evaporation and condensation inside the tube. The reaction in the dark is desirable to avoid fluorescence quenching, but not mandatory.
- Use a natural convection or forced air flow convection oven, or PCR device. The reaction is stopped by adding 3 μl of 5x SDS-PAGE sample loading buffer to each tube, and heating it at 95 °C for 1 min.
- Centrifuge the tubes briefly to spin down the contents. Optionally, EDTA (ethylenediaminetetraacetic acid) is added at a final concentration of 5-10 mM, before the addition of the SDS-PAGE sample loading buffer, to stop the reaction immediately. Note that placing the tubes (in an aluminum block) on ice is sometimes inadequate to stop the reaction completely.
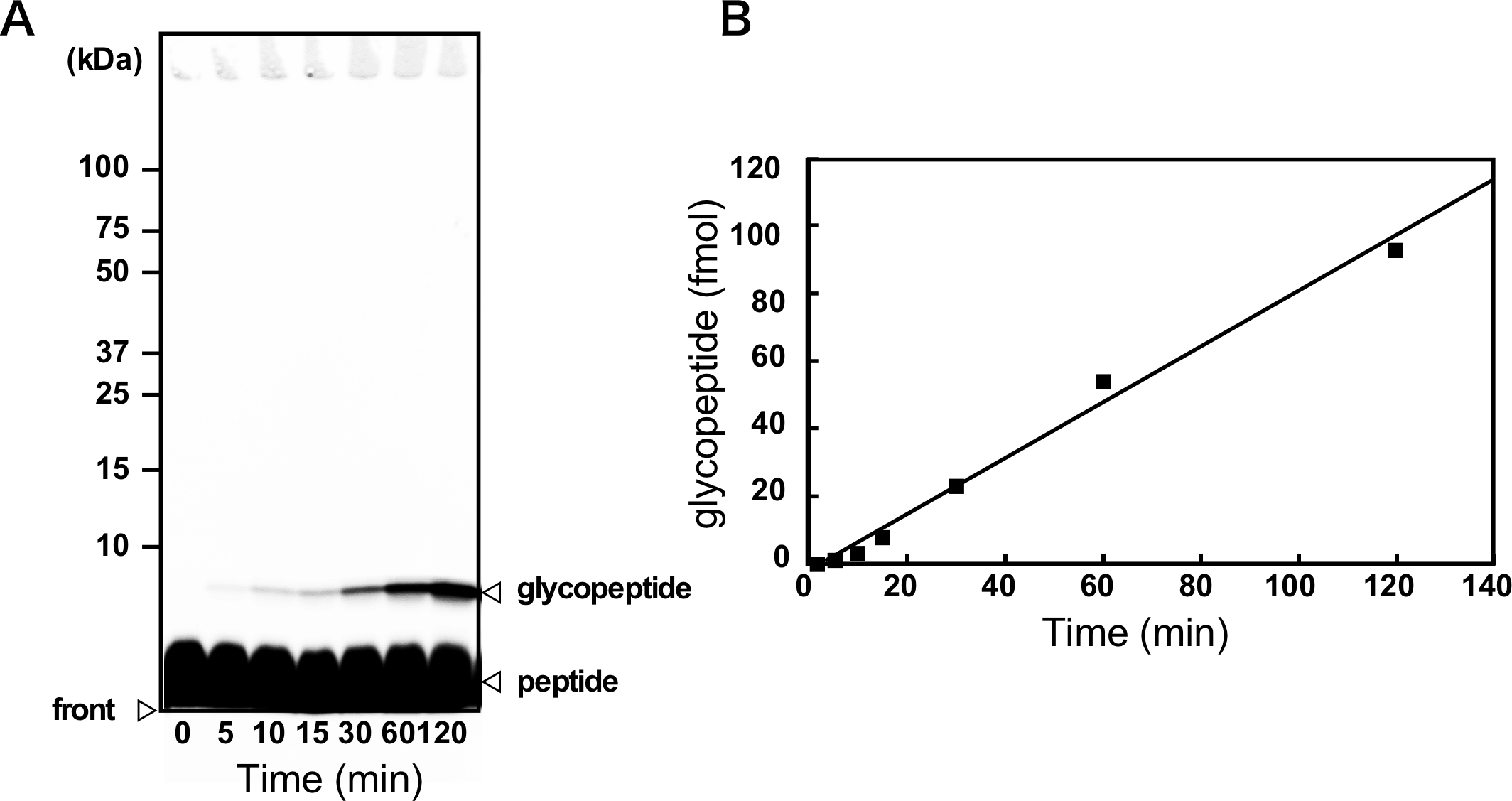
Figure 3. Time-course of glycopeptide production. A. Glycopeptide products were separated from unmodified peptides by SDS-PAGE and quantified by fluorescence imaging. The fluorescent dye is attached to the peptide. The positions of the protein standard and the dye front are indicated. The contrast was adjusted with a sigmoid tone curve. This gel image is adapted from (Matsumoto et al., 2013). B. The change in the amount of the glycopeptide product with time should produce a straight line for quantitative measurement.
- Preparation of reaction buffer solution
- Electrophoresis
The heating in the presence of SDS is not necessary for PAGE separation of (glyco)peptides, but may be useful to clarify the migration image by its denaturing effects on miscellaneous proteins in the reaction mixture solution.
Load the whole volume (15 μl) of the samples on the SDS-PAGE gels. If the sample volume decreases for any reason, a small volume of water or buffer may be added, but keep in mind that the entire sample must be loaded in one of the gel wells. Perform electrophoresis at about 23 mA per gel in the constant current mode for about 75 min. The optimal constant current is lower than the standard current for the separation of polypeptides, 30-40 mA. The appropriate setting of the electric current is essential for good separation of glycopeptide products from unreacted peptides. As a guideline, the time of electrophoresis is set for the bromphenol blue (BPB) front line to reach about 1 cm above the bottom of the gel. The run out of gel should be avoided because unreacted peptides will migrate close to the dye front line. The concurrent running of prestained molecular weight markers on the gel is not recommended, because some of the staining dyes are fluorescent.
Use handcasted polyacrylamide gels with a 4% acrylamide(AA)/bisacrylamide(bis) stacking gel and a 20% AA/bis resolving gel, or precast polyacrylamide gels with a 4% AA/bis stacking gel and a 15%-25% gradient AA/bis resolving gel. The high AA/bis percentage is important for the separation between glycopeptides and peptides. In general, precast gels are preferable to handcasted gels, because of the more homogeneous quality of the gel. Gels sandwiched between glass plates should be used, because plastic plates make the fluorescent imaging difficult due to light absorption.
Laemmli-SDS-PAGE based on the Tris-Glycine buffer system is usually used. Alternatively, Tricine-SDS-PAGE based on the Tris-Tricine buffer system is effective for better separation of glycopeptides. In fact, a peptide with just one monosaccharide residue can be resolved from the unmodified peptide (Gerber et al., 2013). One should take precautions, however, the mobility of glycopeptides could be affected by sugar composition and branching pattern. - Detection by in-gel fluorescent imaging
(Glyco)peptides are visualized in-gel using a fluorescence imager. It is unnecessary to detach the glass plates from the gel.- Immediately after the electrophoresis finishes, clean the glass plates with water and then with ethanol.
- Blow off the residual ethanol with an air (gas) duster. Do not use paper towels or cloths, because traces of the wiping will remain on the glass plates.
- Set up the fluorescence imager as described in the manual. In the case of the LAS-3000 multicolor image analyzer (Fuji Film) with the TAMRA fluorophore, use Precision mode, Standard sensitivity, green LED (Green 520 nm EPI) illuminator, a 575DF20 filter, and Iris 0.85. The typical exposure time is 1 to 60 s. In principle, the exposure time must be optimized for any glycosylated, and non-glycosylated bands should not be overexposed for the accurate quantification of the bands.
- Possible negative controls include the omission of the detergent-solubilized OST or LLO in the reaction mixture. Instead, peptides containing non-functional sequons, NPT or NPS, can be used. By contrast, the preparation of a positive control is not easy, since the chemical synthesis of glycopeptides are difficult and expensive. Alternatively, after gel extraction from the product bands, mass analysis using MALDI-TOF-MS or ESI-MS is the best way to identify the glycosylation of the substrate peptides.
Data analysis
A typical fluorescent image of an SDS-PAGE gel is shown in Figure 4. The magenta color indicates the saturation of the fluorescent intensity. A peptide library experiment with low efficient sequons requires a long exposure time to detect the glycopeptide product bands. Note the small variation in the migration rates of peptides and glycopeptides with different amino acid sequences during electrophoresis.
Select the band boundary of a (glyco)peptide to obtain the fluorescence intensity, and in parallel, select a blank boundary with the same area for subtraction of the background noise. The ImageGauge software (Fuji Film) has a special freehand tool to set band boundaries and blank boundaries with a free curve (Figure 4C).
For the determination of the absolute quantity of (glyco)peptides by fluorescent imaging, a standard calibration curve can be made, using any peptide with the TAMRA fluorophore. Caution must be exercised when preparing serial dilutions of the TAMRA-peptide to avoid adsorption on plastic tubes: it should be mixed with the SDS-PAGE sample loading buffer first, and then diluted. Alternatively, the addition of small amount of Tween 20 (0.02%) is effective to prevent adsorption on plastic tubes.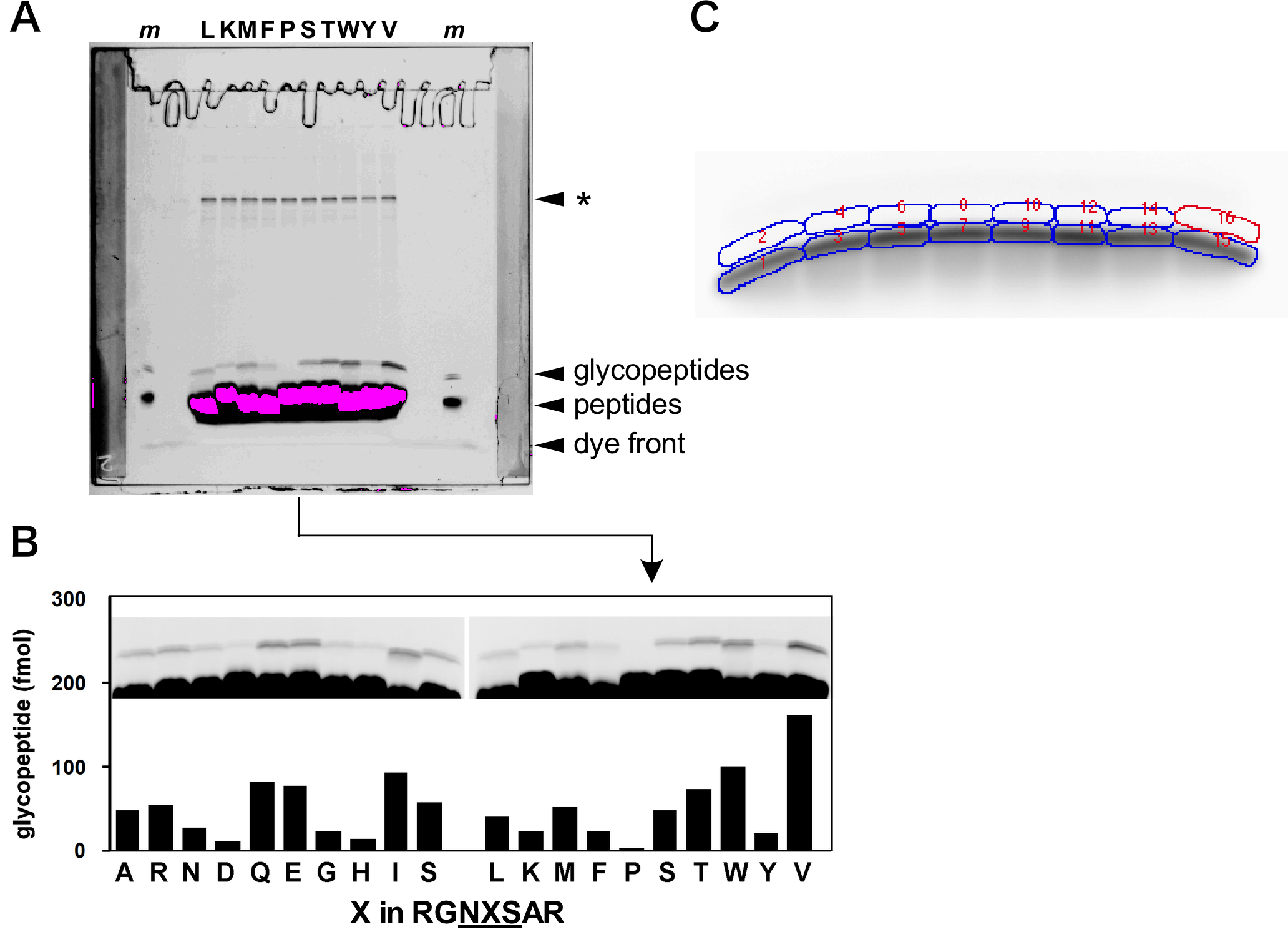
Figure 4. Effects of the amino acid substitutions at the second position of the N-glycosylation sequon on the efficiency of the oligosaccharide transfer reaction. A. The original fluorescent image of the peptide library experiment performed in (Igura and Kohda, 2011a). The asterisk shows the unidentified bands, which only appear when the TAMRA fluorophore is attached to the peptides through the thiol-maleimide reaction. When the TAMRA fluorophore is directly attached to the N-terminal amino group through amide bond formation, no extra bands appear (as shown in lanes marked with m). B. Part of Figure 1A in (Igura and Kohda, 2011a) is redisplayed for reference. C. For background correction, band boundaries and blank boundaries were set by the Freehand Tool option of the ImageGauge software (Fuji Film). The contrast was adjusted with a sigmoid tone curve.
Notes
- Oligosaccharyltransferase
The animal, plant, and fungal OSTs are multi-subunit protein complexes consisting of eight different membrane subunit proteins (Larkin and Imperiali, 2011). In contrast, the lower eukaryotic protozoan OST is a single-subunit enzyme consisting solely of the catalytic subunit, and is referred to as Stt3. The archaeal and eubacterial OSTs are also single-subunit enzymes, consisting of AglB and PglB, respectively. Even though the catalytic subunits have different names, they have a common ancestor and share several conserved short amino-acid sequence motifs. Each genome usually encodes multiple paralogous genes of the catalytic subunit (and other subunits). Thus, it is better to prepare a recombinant OST protein using appropriate host cells. For example, the OST proteins from Pyrococcus (Igura and Kohda, 2011b), Archaeoglobus (Matsumoto et al., 2013), and Campylobacter (Maita et al., 2010; Gerber et al., 2013) were successfully expressed in the membrane fractions of E. coli cells. Exceptionally, the yeast genome encodes only one Stt3 gene. The yeast OST complex (containing either the Ost3 or Ost6 subunit) can be prepared from yeast cells (Harada et al., 2013). Alternatively, one paralogous OST protein from crude membrane fractions of a natural source, for example Pyrococcus furiosus, can be purified by polyclonal antibodies (Igura et al., 2008). The polyclonal antibodies can be obtained by immunization of rabbits with a fragment corresponding to the C-terminal soluble domain expressed in E. coli cells.
To prepare a large amount of a glycopeptide product, the crude membrane fractions, which contain both wild-type OST and LLO, prepared from any cultured cells are usable. The detergent solubilization of the crude membrane fractions is effective to improve the storage life of the membrane fractions. Note that the choice of detergent is critical for LLO solubilization. For the preparation of yeast and archaeal membrane fractions, Triton X-100 is preferable to digitonin or DDM for the co-solubilization of OST and LLO. The incubation with an acceptor peptide readily produces glycopeptide products. Serious side reactions, such as O-glycosylation, do not occur since the water-soluble compounds, such as nucleotide sugars, are washed away from the membrane fractions. One side reaction we encountered is the deamination of the C-terminal α-amide group. The addition of protease inhibitors is recommended, even though the oligosaccharide chains on asparagine effectively protect the glycopeptide from protease digestion. Incorporation of a biotin group during peptide synthesis is helpful for the purification of large quantities of glycopeptides (Fujinami et al., 2017). - Sugar donor LLO
Crude LLO preparations were described for yeast (Kelleher et al., 2001), protists (Samuelson et al., 2005), Pyrococcus (Igura et al., 2008), Archaeoglobus (Matsumoto et al., 2013), Haloferax (Guan et al., 2010), and Campylobacter (Maita et al., 2010; Gerber et al., 2013). Briefly, cultured cells harvested by centrifugation are disrupted by glass beads or sonication, and cell debris is removed by centrifugation. Optionally, the supernatant is ultracentrifuged at 100,000 x g, and the membrane pellet is resuspended in buffer. The two-phase partitioning of a chloroform:methanol:water system produces the crude LLO fractions. The extracted crude LLOs can be further purified by anion exchange chromatography in a chloroform:methanol:water solvent system, and subsequently by NP-HPLC (normal phase high-performance liquid chromatography) using a chloroform:methanol:water:NH4OH solvent system (Guan et al., 2010; Taguchi et al., 2016). The crude and purified LLO solubilized in chloroform:methanol:water is stored in a glass container at -20 °C until use. - Sugar acceptor peptide
- Design of sugar acceptor peptides
The sugar acceptor peptide is usually prepared by custom peptide synthesis. Typically, it consists of three to ten amino acid residues containing an N-glycosylation sequon, Asn-X-Thr or Asn-X-Ser (X ≠ Pro). Just three residues corresponding to the N-glycosylation sequon can serve as a substrate, if the N-terminal α-amino group and C-terminal α-carboxy group are both blocked by an acetyl group and an amide group, respectively. In the original report (Kohda et al., 2007), (TAMRA)-Gly-Asn-Ser-Thr-Val-Thr-NH2 was used, where the underlined sequence is the N-glycosylation sequon, for the oligosaccharyl transfer assays of the yeast and Pyrococcus OSTs. The underlined sequence was changed to Gln-Ser-Thr, Asn-Pro-Thr, or Asn-Ser-Ala to test the N-glycosylation sequon as negative controls. The eubacterial Campylobacter N-glycosylation sequon requires an acidic residue, Asp or Glu, at the -2 position. (TAMRA)-Ala-Asp-Gln-Asn-Ala-Thr-Tyr-Lys-COOH and (5-CF)-Gly-Ser-Asp-Gln-Asn-Ala-Thr-Phe-NH2 were used for the Campylobacter OSTs (Maita et al., 2010; Gerber et al., 2013). The 5-CF stands for 5-carboxyfluorescein, alias 5-FAM. Considering the solubility in acidic aqueous solutions and the sensitive detection by RP (reverse phase)-HPLC-ESI-MS (electron spray ionization mass spectrometry), the incorporation of lysine and arginine residues is recommended in the peptide sequence. For example, (TAMRA)-Ala-Ala-Tyr-Asn-Val-Thr-Lys-Arg-(Lys-biotin)-COOH was used for the Pyrobaculum OST (Fujinami et al., 2017).
To make the peptides better substrates, Thr is preferable to Ser at the +2 position, and Asp is preferable to Glu at the -2 position. Pro must be avoided at the +1 (X) position and should be avoided at the -1 and +3 positions. The choice of the amino acid residue at the +1 (X) position has a high impact on the oligosaccharyl transfer activity. Several amino acid residues should be tested for selecting the best acceptor peptide. For example, in in vitro assay conditions, Val is the most favored amino acid residue at the +1 position for the Pyrococcus OST (Igura and Kohda, 2011b), Glu and Gln are best for the Archaeoglobus OST (Matsumoto et al., 2017), and Ala, Ser, and positively charged residues are ideal for the Campylobacter OST (Chen et al., 2007). Similarly, experimental analyses of an individual N-glycosylation sequon in a particular protein showed the pronounced effects of amino acid variations at the +1 position on the glycosylation efficiency with the canine OST, in microsomes supplemented with a rabbit reticulocyte cell-free translation system (Shakin-Eshleman et al., 1996; Kasturi et al., 1997). In contrast, the strong preference at the +1 position disappeared when the peptide was cross-linked to the Archaeoglobus OST protein (Matsumoto et al., 2017), and was not detected in comprehensive statistical analyses of the eukaryotic and archaeal glycosylation sites (Ben-Dor et al., 2004; Petrescu et al., 2004; Abu-Qarn and Eichler, 2007).
In the original report, TAMRA (carboxytetramethylrhodamine, absorption/emission, 555 nm/580 nm) was attached to the N-terminal α-amino group during custom peptide synthesis (Kohda et al., 2007). Frequently, the TAMRA fluorophore is a mixture of two isomers, 5- and 6-carboxytetramethylrhodamines, and the 5- and 6-isomer carrying peptides split into two peaks in the RP-HPLC chromatogram. We found that the slowly eluting peak of the TAMRA peptide yielded a clearer SDS-PAGE background in the OST assay. If the background of the fluorescence in-gel image is high, then the other isomer-carrying peptide should be used, to avoid contamination with unwanted fluorescent impurities during the RP-HPLC purification. Instead of the incorporation of TAMRA or another fluorescent dye during peptide synthesis, an additional cysteine residue can be incorporated in the peptide sequence. The sulfhydryl (SH) group of the cysteine residue is useful as an anchoring point for fluorescent labeling by a thiol-maleimide reaction (Figure 4). - Preparation of acceptor peptide solutions
Dissolve lyophilized peptide powder in water. The pH of the peptide solution is very acidic (pH < 3). Neutralize the acidic solution by serial additions of a small volume of alkaline solution (e.g., 1 M NaOH). The use of pH test paper is sufficient. Store the concentrated peptide solution (10 mM) in conveniently sized aliquots at -20 °C or -80 °C. To prepare the working stock solution, dilute to the concentration of the stock to 10-100 μM with 0.02% Tween 20. The working stock solution can be stored at 4 °C in a refrigerator for up to several weeks. However, long-term refrigerated storage results in degradation, and may produce high background levels in the fluorescent images of the SDS-PAGE gels.
The concentration of a TAMRA-peptide is determined by the absorbance at 555 nm, with an extinction coefficient of 90,000 M-1 cm-1 in aqueous solution. For example, 10 μM of the TAMRA fluorophore corresponds to an absorbance of 0.9 at 555 nm. The concentration of a 5-CF (alias 5-FAM)-peptide is determined by the absorbance at 495 nm, with an extinction coefficient of 79,000 M-1 cm-1 in aqueous solution.
- Design of sugar acceptor peptides
Recipes
- 10:10:3 (v/v/v) chloroform:methanol:water
- Measure and mix all solvent in glass containers, since chloroform will dissolve plastic
- Store the solution in a capped glass container to prevent concentration changes due to the evaporation of chloroform and methanol
- 5x SDS-PAGE sample loading buffer
0.25 M Tris-HCl, pH 6.8
5% SDS
50% (w/v) glycerol
0.125% (w/v) bromophenol blue
Store working aliquots at room temperature. The final percentage of SDS should not exceed 1% to avoid band broadening - Running electrophoresis buffer
0.025 M Tris-HCl, pH 8.3
0.192 M glycine
0.1% SDS
Acknowledgments
This work was supported by JSPS KAKENHI Grants JP15083205 and JP26119002 (to DK).
Competing interests
The authors declare there are no potential conflicts of interest.
References
- Abu-Qarn, M. and Eichler, J. (2007). An analysis of amino acid sequences surrounding archaeal glycoprotein sequons. Archaea 2(2): 73-81.
- Bause, E. and Hettkamp, H. (1979). Primary structural requirements for N-glycosylation of peptides in rat liver. FEBS Lett 108(2): 341-344.
- Ben-Dor, S., Esterman, N., Rubin, E. and Sharon, N. (2004). Biases and complex patterns in the residues flanking protein N-glycosylation sites. Glycobiology 14(2): 95-101.
- Chen, M. M., Glover, K. J. and Imperiali, B. (2007). From peptide to protein: comparative analysis of the substrate specificity of N-linked glycosylation in C. jejuni. Biochemistry 46(18): 5579-5585.
- Cherepanova, N., Shrimal, S. and Gilmore, R. (2016). N-linked glycosylation and homeostasis of the endoplasmic reticulum. Curr Opin Cell Biol 41: 57-65.
- Fujinami, D., Taguchi, Y. and Kohda, D. (2017). Asn-linked oligosaccharide chain of a crenarchaeon, Pyrobaculum calidifontis, is reminiscent of the eukaryotic high-mannose-type glycan. Glycobiology 27(8): 701-712.
- Geetha-Habib, M., Park, H. R. and Lennarz, W. J. (1990). In vivo N-glycosylation and fate of Asn-X-Ser/Thr tripeptides. J Biol Chem 265(23): 13655-13660.
- Gerber, S., Lizak, C., Michaud, G., Bucher, M., Darbre, T., Aebi, M., Reymond, J. L. and Locher, K. P. (2013). Mechanism of bacterial oligosaccharyltransferase: in vitro quantification of sequon binding and catalysis. J Biol Chem 288(13): 8849-8861.
- Guan, Z., Naparstek, S., Kaminski, L., Konrad, Z. and Eichler, J. (2010). Distinct glycan-charged phosphodolichol carriers are required for the assembly of the pentasaccharide N-linked to the Haloferax volcanii S-layer glycoprotein. Mol Microbiol 78(5): 1294-1303.
- Harada, Y., Buser, R., Ngwa, E. M., Hirayama, H., Aebi, M. and Suzuki, T. (2013). Eukaryotic oligosaccharyltransferase generates free oligosaccharides during N-glycosylation. J Biol Chem 288(45): 32673-32684.
- Igura, M. and Kohda, D. (2011a). Quantitative assessment of the preferences for the amino acid residues flanking archaeal N-linked glycosylation sites. Glycobiology 21(5): 575-583
- Igura, M. and Kohda, D. (2011b). Selective control of oligosaccharide transfer efficiency for the N-glycosylation sequon by a point mutation in oligosaccharyltransferase. J Biol Chem 286(15): 13255-13260.
- Igura, M., Maita, N., Kamishikiryo, J., Yamada, M., Obita, T., Maenaka, K. and Kohda, D. (2008). Structure-guided identification of a new catalytic motif of oligosaccharyltransferase. EMBO J 27(1): 234-243.
- Kasturi, L., Chen, H. and Shakin-Eshleman, S. H. (1997). Regulation of N-linked core glycosylation: use of a site-directed mutagenesis approach to identify Asn-Xaa-Ser/Thr sequons that are poor oligosaccharide acceptors. Biochem J 323 (Pt 2): 415-419.
- Kelleher, D. J., Karaoglu, D. and Gilmore, R. (2001). Large-scale isolation of dolichol-linked oligosaccharides with homogeneous oligosaccharide structures: determination of steady-state dolichol-linked oligosaccharide compositions. Glycobiology 11(4): 321-333.
- Kohda, D., Yamada, M., Igura, M., Kamishikiryo, J. and Maenaka, K. (2007). New oligosaccharyltransferase assay method. Glycobiology 17(11): 1175-1182.
- Larkin, A. and Imperiali, B. (2011). The expanding horizons of asparagine-linked glycosylation. Biochemistry 50(21): 4411-4426.
- Maita, N., Nyirenda, J., Igura, M., Kamishikiryo, J. and Kohda, D. (2010). Comparative structural biology of eubacterial and archaeal oligosaccharyltransferases. J Biol Chem 285(7): 4941-4950.
- Matsumoto, S., Shimada, A., Nyirenda, J., Igura, M., Kawano, Y. and Kohda, D. (2013). Crystal structures of an archaeal oligosaccharyltransferase provide insights into the catalytic cycle of N-linked protein glycosylation. Proc Natl Acad Sci U S A 110(44): 17868-17873.
- Matsumoto, S., Taguchi, Y., Shimada, A., Igura, M. and Kohda, D. (2017). Tethering an N-glycosylation sequon-containing peptide creates a catalytically competent oligosaccharyltransferase complex. Biochemistry 56(4): 602-611.
- Petrescu, A. J., Milac, A. L., Petrescu, S. M., Dwek, R. A. and Wormald, M. R. (2004). Statistical analysis of the protein environment of N-glycosylation sites: implications for occupancy, structure, and folding. Glycobiology 14(2): 103-114
- Samuelson, J., Banerjee, S., Magnelli, P., Cui, J., Kelleher, D. J., Gilmore, R. and Robbins, P. W. (2005). The diversity of dolichol-linked precursors to Asn-linked glycans likely results from secondary loss of sets of glycosyltransferases. Proc Natl Acad Sci U S A 102(5): 1548-1553.
- Schwarz, F., Lizak, C., Fan, Y. Y., Fleurkens, S., Kowarik, M. and Aebi, M. (2011). Relaxed acceptor site specificity of bacterial oligosaccharyltransferase in vivo. Glycobiology 21(1): 45-54.
- Shakin-Eshleman, S. H., Spitalnik, S. L. and Kasturi, L. (1996). The amino acid at the X position of an Asn-X-Ser sequon is an important determinant of N-linked core-glycosylation efficiency. J Biol Chem 271(11): 6363-6366.
- Sharma, C. B., Lehle, L. and Tanner, W. (1981). N-Glycosylation of yeast proteins. Characterization of the solubilized oligosaccharyl transferase. Eur J Biochem 116(1): 101-108.
- Taguchi, Y., Fujinami, D. and Kohda, D. (2016). Comparative analysis of archaeal lipid-linked oligosaccharides that serve as oligosaccharide donors for asn glycosylation. J Biol Chem 291(21): 11042-11054.
- Wacker, M., Linton, D., Hitchen, P. G., Nita-Lazar, M., Haslam, S. M., North, S. J., Panico, M., Morris, H. R., Dell, A., Wren, B. W. and Aebi, M. (2002). N-linked glycosylation in Campylobacter jejuni and its functional transfer into E. coli. Science 298(5599): 1790-1793.
Article Information
Copyright
© 2019 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Yamasaki, T. and Kohda, D. (2019). A Radioisotope-free Oligosaccharyltransferase Assay Method. Bio-protocol 9(5): e3186. DOI: 10.21769/BioProtoc.3186.
Category
Biochemistry > Carbohydrate > Oligosaccharide
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










