- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Detection of Disulfides in Protein Extracts of Arabidopsis thaliana Using Monobromobimane (mBB)
Published: Vol 9, Iss 5, Mar 5, 2019 DOI: 10.21769/BioProtoc.3183 Views: 6423
Reviewed by: Dennis J NürnbergCiaran McFarlaneAgnieszka Zienkiewicz

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Histone Deubiquitination Assay in Nicotiana benthamiana
Shujing Liu and Lars Hennig
Mar 5, 2018 6693 Views

Simple Method to Determine Protein Redox State in Arabidopsis thaliana
Keisuke Yoshida and Toru Hisabori
Jun 5, 2019 7477 Views

In vitro Auto- and Substrate-Ubiquitination Assays
Hye Lin Park [...] Gyeong Mee Yoon
Apr 5, 2022 3546 Views
Abstract
Thiol-disulfide exchange is a key posttranslational modification, determining the folding process of intra- and inter-protein structures. Thiols can be detected by colorimetric reagents, which are stoichiometrically reduced by free thiols, and by fluorescent adducts, showing fluorescence only after thioester formation. We adapted a simple three-step method for detection of disulfide bonds in proteins. After irreversible blocking of protein thiols, disulfide bonds are reduced, followed by the detection of thiols. The approach presented here provides an economical procedure that can be used to obtain a global overview of the thiol-disulfide status of proteins in plants. This method allows the detection of modifications in samples on a gel and can be used for semi-quantitative analysis.
Keywords: Protein labelingBackground
The redox status of proteins plays a key role in a number of cellular processes. Thiol-disulfide exchange reactions support the redox control of plant responses to fluctuating light under natural environments via a photochemical electron transfer cascade for posttranslational modification of proteins. Whereas thiols (R-SH) have relatively higher reactivity than other cellular components and can be directly detected by a variety of reagents and separation techniques (Fahey et al., 1980 and 1981; Newton et al., 1981; Riddles et al., 1983; Fenton and Fahey, 1986; Tyagarajan et al., 2003), disulfide bonds (R-S-S-R) have no such chemical characteristics. Therefore, the most common methodology for the detection of disulfide bonds consists of a three-step procedure: blocking of thiols, reduction of disulfide bonds, and subsequent detection of additionally exposed thiols. Dithiothreitol (DTT) is a well-known and frequently used reducing agent for the protection and exposure of thiol groups of proteins. However, the use of DTT in the laboratory is undesirable because of its harmful odor. Moreover, it can subsequently interfere with thiol detection reagents unless it is appropriately removed (Getz et al., 1999). Also, the reducing power of DTT is limited to pH > 7. Tris(2-carboxyethyl) phosphine-hydrochloride (TCEP) can be a promising alternative with advantages of being odorless and more effective in the reduction of disulfide bonds over a wide pH range from 1.5 to 8.5. Monobromobimane (mBB) has been used for fluorescent labeling of low molecular weight thiols by detecting fluorescent derivatives under a fluorescent microscope or with high-performance liquid chromatography (Fahey and Newton, 1987; Meyer et al., 2001). This protocol describes a simple method for discriminate labeling with mBB of free thiols, disulfide bonds, and total thiols in plant protein extracts. Here, we show an example of the procedure using Arabidopsis leaves. Free thiols are labeled without any preprocessing (Figure 1A), whereas labeling of disulfide bonds requires blocking of free thiols by iodoacetamide (IAA) and reduction of disulfide bonds by TCEP before the mBB treatment (Figure 1B). Total labeling of thiols just after TCEP treatment is helpful to estimate the redox state of total thiols in protein extracts (Figure 1C). This simple method allows the detection of modifications in samples on a gel and can be used for semi-quantitative analysis and provides an overview of the redox state of thiol-containing proteins without any special equipment other than conventional protein electrophoresis and imaging systems.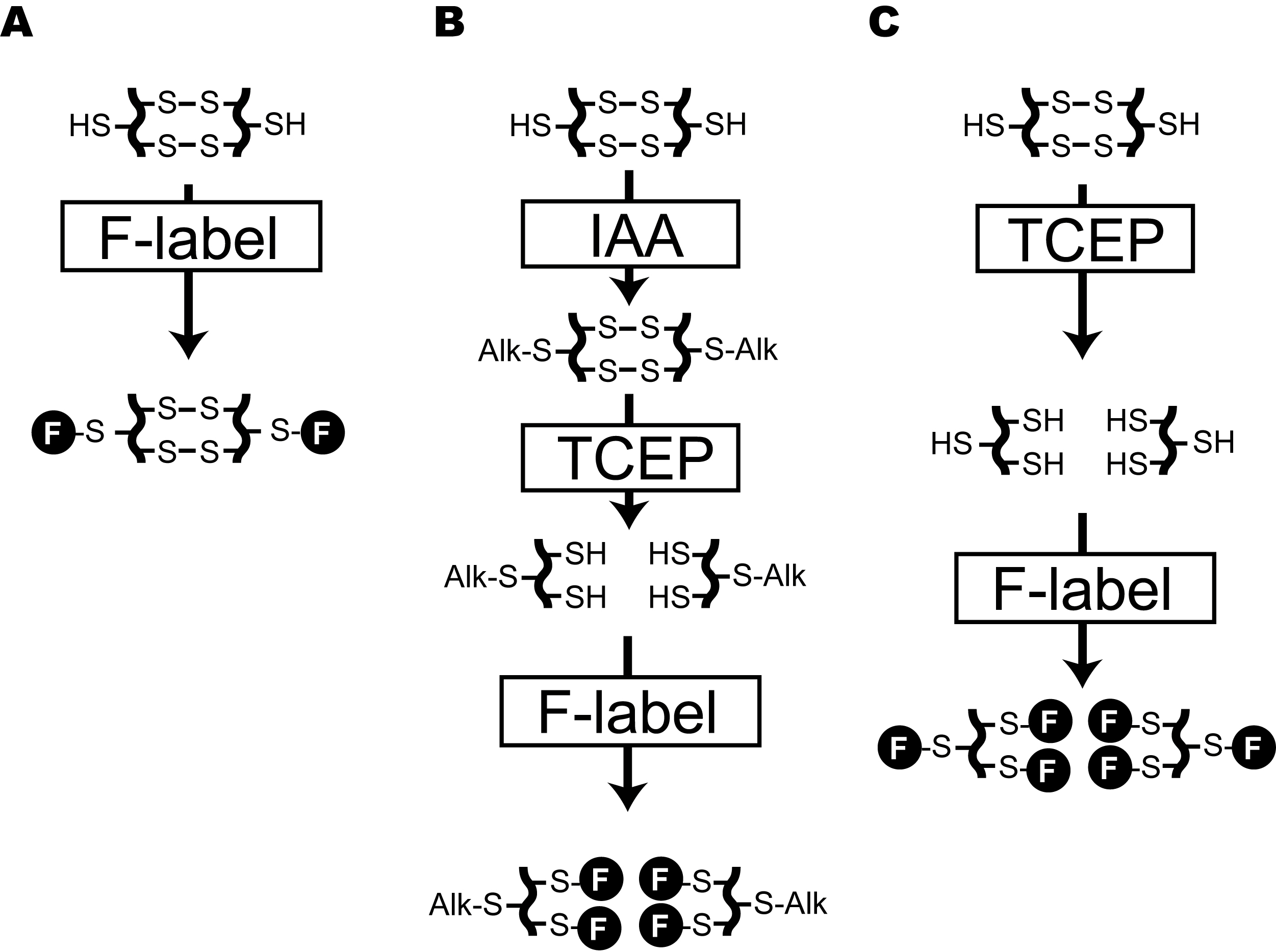
Figure 1. Flowchart for discriminate labeling of thiols in protein under different redox states. A. Labeling of free thiols in proteins. B. Specific labeling of thiols, forming disulfide bonds in protein. C. Total labeling of thiols in proteins. F in closed circle means mBB-derived fluorescence. IAA: Iodoacetamide, TCEP: Tris(2-carboxyethyl)phosphine Hydrochloride.
Materials and Reagents
- Finntip Pipette Tips (Finntip Flex 10, 200, and 1,000)
- 1.5 ml microcentrifuge tube (VIOLAMO, F-1.5, catalog number: 1-1600-01)
- Arabidopsis thaliana wild-type plants (Ecotype: Col-0)
- Arabidopsis thaliana mutant plant of interest (Ecotype: Col-0)
- Liquid nitrogen
- Tris(hydroxymethyl)aminomethane (Tris) (Nacalai Tesque, catalog number: GR35406-91)
- cOmpleteTM, Mini, Protease Inhibitor Cocktail (Roche, catalog number: 4693159001)
- Bio-Rad Protein Assay Kit (Bio-Rad, catalog number: 5000001ja)
- FOCUSTM Protein Alkylation (TaKaRa, catalog number: GA503)
- Iodoacetamide (IAA) (Sigma-Aldrich, catalog number: I1149-5G)
- Trichloroacetic acid solution (TCA), 100% (w/v) (Nacalai Tesque, catalog number: SP06275-24)
- Acetone (Nacalai Tesque, catalog number: SP09888-85)
- FOCUSTM Protein Reductant (TaKaRa, catalog number: GA501)
- Tris(2-carboxyethyl)phosphine Hydrochloride (TCEP) (Nacalai Tesque, catalog number: SP06342-21)
- Monobromobimane (mBB) (Tokyo Chemical Industry, catalog number: B4220)
- Glycerol (Nacalai Tesque, catalog number: SP17045-94)
- Bromophenol Blue (BPB) (Nacalai Tesque, catalog number: B-2609)
- Glycine (Nacalai Tesque, catalog number: SP17141-24)
- Sodium lauryl sulfate (SDS) (Nacalai Tesque, catalog number: SP08933-34)
- Protein extraction buffer (see Recipes)
- IAA solution (see Recipes)
- SDS-PAGE sample buffer (see Recipes)
- SDS-PAGE buffer (see Recipes)
Equipment
- Pipettes (Thermo Fisher Scientific, FinnpipetteTM F2)
- Cork borer (4.0 mm, inside diameter of the hole)
- Conventional Dewar’s vessel
- Vortexer (Scientific Industries, Vortex-Genie 2)
- Homogenizer pestle (VIOLAMO, F-1.5, catalog number:1-2955-02)
- Refrigerated microcentrifuge (TOMY, MX305)
- Absorption spectrometer (Thermo Fisher Scientific, MultiskanTM GO)
- Mini-PROTEAN Tetra System (Bio-Rad, catalog number: 1658004ja)
- Mini-PROTEAN TGX Gels (Bio-Rad, catalog number: 456-1095)
- Power supply (ATTO, catalog number: AE-8135)
- UV Transilluminator (Maestrogen, model: UV-01)
- Yellow filter (Kenko, Black & White Filter, Y2 Professional, λcut-on 470-480 nm)
- Conventional digital camera with hood (RICHO, WG-4)
Software
- ImageJ (https://imagej.nih.gov/ij//)
Procedure
- Protein extraction
- Place freshly prepared leaf discs (10-50 mg) punched-out with a cork borer in a 1.5 ml microcentrifuge tube and immediately freeze in liquid nitrogen in a Dewar’s vessel. The estimated average weight of five leaf discs should be approximately 10 mg.
- Grind the sample in the microcentrifuge tube with a homogenizer pestle and add 200 µl of the ice-cold protein extraction buffer (prepare immediately prior to use, Recipe 1) per 50 mg sample on ice before sample lysis.
Note: Make sure not to dissolve the sample. We recommend grinding the sample in the presence of liquid nitrogen rather than on ice. In the case of Arabidopsis leaf discs, it is easy to obtain fully homogenized sample in a couple of seconds. - Centrifuge at 14,000 x g, 4 °C for 10 min.
- Transfer the supernatant to a new 1.5 ml microcentrifuge tube for removing sample debris.
- Protein measurement
- Prepare the dye for Bio-Rad Protein Assay by diluting 1 volume of the dye reagent concentrate with 4 volumes of distilled, deionized (DDI) water.
- Add 2 and 5 µl protein extract into 198 and 195 µl diluted dye reagent, respectively.
- After 5 min, measure absorbance at 595 nm and estimate the protein concentration with protein standard II (BSA) in Bio-Rad Protein Assay Kit (0.2 to 2.0 mg/ml).
- Thiol alkylation
- Adjust the protein concentration to 1.0 mg/ml with the extraction buffer and combine several samples to prepare 1.5 ml of protein extract in a single tube.
- Divide the solution into three 1.5 ml microcentrifuge tubes. Each tube contains 500 µl of the extract, one each for the determination of free thiols (tube A), disulfide bonds (tube B), and total thiols (tube C) in the protein extracts. All the tubes should be kept on ice until use.
Note: For all experiments, we strongly recommend starting from a single protein extract to discriminately detect free thiols, disulfide bonds, and total thiols in proteins to minimize the sample variations. - Add 2.5 µl FOCUSTM alkylation buffer to tubes B and C, and vortex for 10 s.
- Add 25 µl 0.4 M IAA (prepare immediately prior to use, Recipe 2) only in tube B for detection of disulfide bonds.
- Incubate tubes B and C without shaking at room temperature (approximately 23 °C), under dark for 30 min.
- Protein purification
- Add an equal volume of cold 20% (w/v) TCA solution to tubes B and C.
- Centrifuge the tubes at 14,000 x g, 4 °C for 30 min.
- Remove the supernatant, leaving the protein pellet intact.
- Remove TCA by washing pellet twice with 500 µl cold acetone and retain the pellet.
- Disulfide reduction
- Adjust the protein reduction solution by adding 2.5 µl FOCUSTM Reductant buffer and 10 µl FOCUSTM Protein Reductant (or 250 mM TCEP in DDI water) to 487.5 µl DDI water.
- Add 50 µl reduction solution to tubes B and C, and then suspend the pellet by gently pipetting up and down. Here, the pellet is not completely dissolved.
- Incubate the tubes at 55 °C for 1 h. Vortex for 10 s every 10 min and confirm the complete dissolution of pellet after incubation.
- mBB labeling
- Add 25 µl of 60 mM mBB solution to tubes A, B, and C.
Note: TCEP can be involved in dehalogenation of mBB and might weaken the efficiency of fluorescent labeling in solution (Graham et al., 2003). Be sure to keep the mBB concentration higher than that of TCEP in the labeling solution. - Incubate at room temperature (approximately 23 °C) for 15 min.
- To remove excess unreacted mBB, add an equal volume of cold 20% (w/v) TCA solution to tubes A, B, and C.
- Centrifuge the tubes at 14,000 x g, 4 °C for 30 min.
- Remove the supernatant, leaving the protein pellet intact.
- Remove TCA by washing pellet twice each with 500 µl cold acetone and retain the pellet.
- Add 25 µl of 60 mM mBB solution to tubes A, B, and C.
- Separation of the labeled proteins
- Suspend the pellets in 10-25 µl SDS-PAGE sample buffer (can be stored at room temperature for 1 week, Recipe 3) and boil for 3 min.
- Assemble the Mini-PROTEAN TGX gel in Mini-PROTEAN Tetra system and pour SDS-PAGE buffer (can be stored at room temperature for 1 month, Recipe 4).
- Load the boiled samples, equivalent to 20-40 µg of proteins, to the sample well.
Note: The initial amount of protein per tube is approximately 50 µg. Adjust and determine the loading volume based on some pilot experiments in the laboratory. We recommend setting an experimental replicate for SDS-PAGE with approximately 20 µg protein, for example, a replicate with 10 µl of the solution obtained by resuspending the pellet in 25 µl SDS-PAGE sample buffer. - Start the electrophoretic run at 100 V and stop after the samples have traversed the desired distance.
- Protein visualization
- Place the gel on UV transilluminator and irradiate the gel.
- Use yellow filter to obtain fluorescence derived from mBB-labeled protein.
Note: mBB has an excitation maximum at 394 nm and an emission maximum at 490 nm.
Data analysis
- Interpretation of the difference between wild type (Col-0) and mutant is based on densitometric analysis of bands with conventional monochrome photographs or fluorescence images using any conventional software, such as ImageJ (https://imagej.nih.gov/ij//).
- The representative data indicate comparable band intensity in R-SH staining between Col-0 and the mutant (Figure 2). However, only on the basis of R-SH data, we cannot assess the redox state of thiol-containing proteins. Here, the R-S-S-R data show a slight increase in band intensity in the mutant and the “Total” data show a slight decrease in band intensity (Figure 2), suggesting that the mutant has an oxidizing atmosphere for thiol-containing proteins. The combined interpretation clearly raises the possibility that the mutant has an oxidizing atmosphere in thiol-containing proteins and the redox state may contribute to the decrease in the amount of thiol-containing proteins.
- Figure 3 exemplifies several different cases to be assumed. Case A indicates comparable band intensities in R-SH staining between Col-0 and the mutant. Case B indicates the R-SH data, showing a decrease in band intensity in the mutant and Case C indicates the R-SH data, showing an increase in band intensity in the mutant.
- In all these cases, interpretations about the redox state can be changed by the R-S-S-R data and the “Total” data (compare cases A1 to A2, B1 to B2, and C1 to C2).
- For example, in Case B1, the R-S-S-R data demonstrating an increase in band intensity in the mutant support the interpretation based on the R-SH data, suggesting an oxidizing atmosphere in the mutant. On the other hand, in Case B2, the R-S-S-H data demonstrating a decrease in band intensity in the mutant is contrary to the hypothesis based on the R-SH data. The total (R-SH and R-S-S-R) data represent that the mutant has lesser variety of thiol-containing proteins. The combined interpretation raises the possibility that the mutant has a comparable redox state in thiol-containing proteins as Col-0 and, thereby, the free thiol might decrease in Case B2.
- Many Arabidopsis mutants have a variety of proteome, resulting from comprehensive proteomic reorganization in response to mutations in various regulatory networks (Figure 3, Case B2 and C1). To minimize possible misinterpretations regarding the redox state of thiol-containing proteins, it is desirable to obtain not only the R-SH data, but also the R-S-S-R and total thiol data.
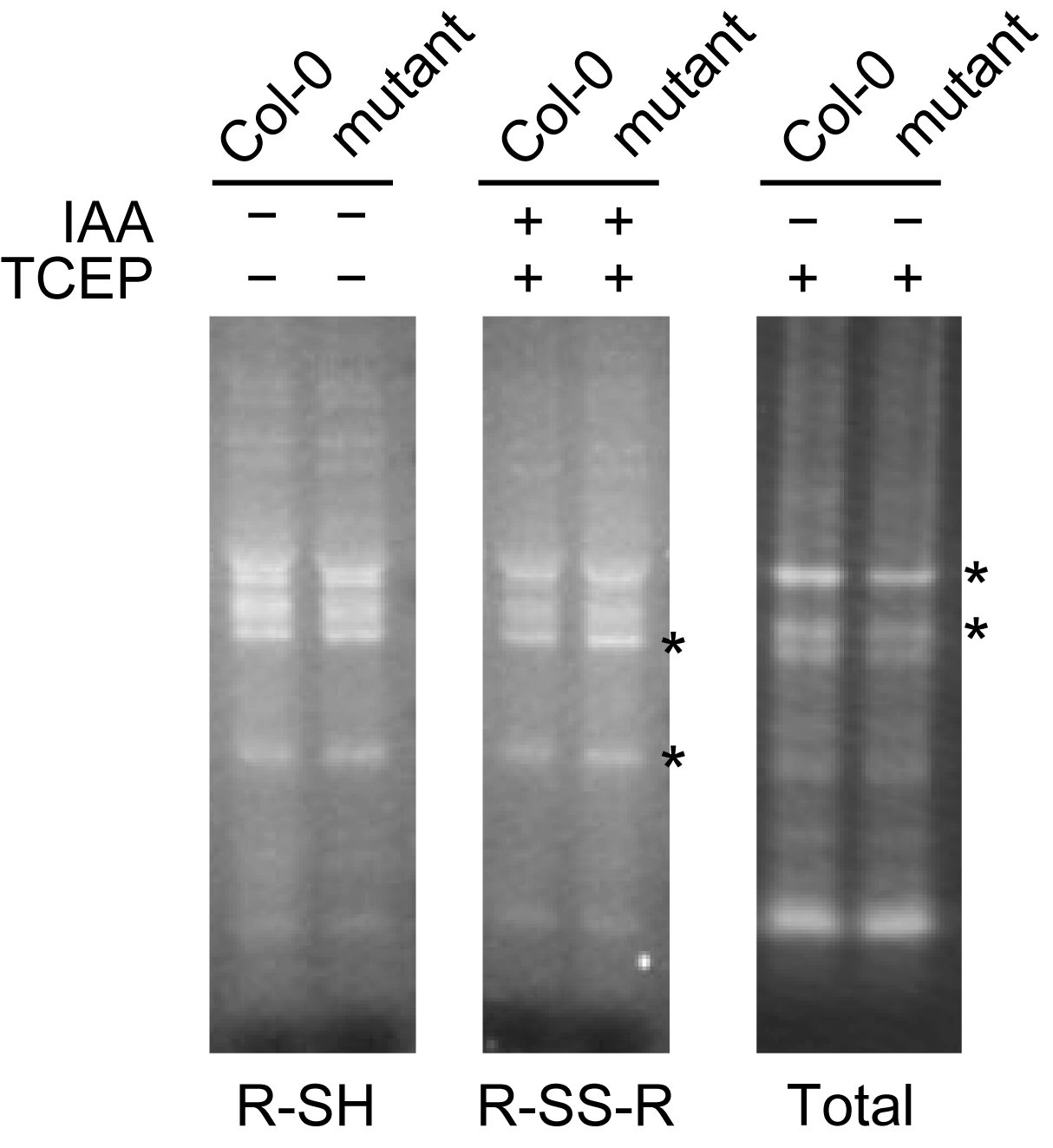
Figure 2. Representative data for monobromobimane (mBB) staining of thiol-containing proteins. Free thiols (R-SH) in proteins were detected by mBB staining without alkylation or reduction (IAA: −, TCEP: −). Disulfide bonds (R-S-S-R) were stained by mBB followed by alkylation and reduction (IAA: +, TCEP: +). Total thiols (R-SH and R-S-S-R) were visualized with mBB only after reduction (IAA: −, TCEP: +). Asterisks indicate different band intensities in Col-0 and the mutant.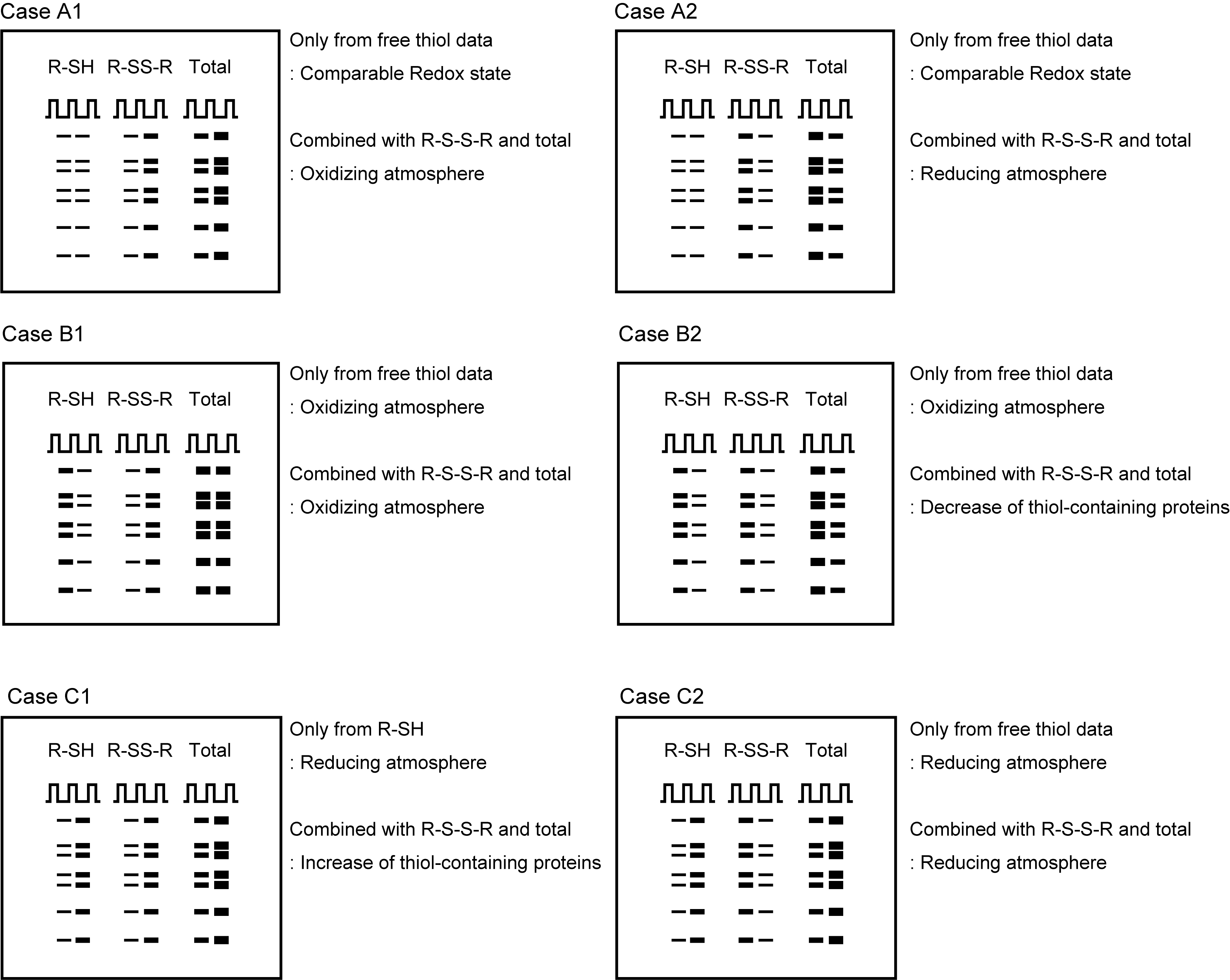
Figure 3. Representative interpretation for the mutant of interest. Representative images of monobromobimane (mBB) stained gels. In all the cases, the left lane is of wild type (Col-0) and the right lane is of the mutant. R-SH and R-S-S-R indicate free thiols and disulfide bonds, respectively, in the proteins.
Recipes
- Protein extraction buffer
30 mM Tris/HCl, pH 7.0, with one tablet of cOmpleteTM, Mini Protease Inhibitor Cocktail per 10 ml - IAA solution
0.4 M IAA - SDS-PAGE sample buffer
50 mM Tris/HCl, pH 7.0
2% (w/v) SDS
5% (w/v) glycerol
0.01% (w/v) BPB - SDS-PAGE buffer
250 mM Tris
1.92 M glycine
1% (w/v) SDS
Acknowledgments
This protocol was adapted and modified from previous work by Hashida et al. (2018). This work was supported by the JSPS KAKENHI Grant Number 17H05714 to M.K.Y.
Competing interests
The authors declare that there are no conflicts of interest.
References
- Fahey, R. C. and Newton, G. L. (1987). Determination of low-molecular-weight thiols using monobromobimane fluorescent labeling and high-performance liquid chromatography. Methods Enzymol 143: 85-96.
- Fahey, R. C., Newton, G. L., Dorian, R. and Kosower, E. M. (1980). Analysis of biological thiols: derivatization with monobromotrimethylammoniobimane and characterization by electrophoresis and chromatography. Anal Biochem 107(1): 1-10.
- Fahey, R. C., Newton, G. L., Dorian, R. and Kosower, E. M. (1981). Analysis of biological thiols: quantitative determination of thiols at the picomole level based upon derivatization with monobromobimanes and separation by cation-exchange chromatography. Anal Biochem 111(2): 357-365.
- Fenton, S. S. and Fahey, R. C. (1986). Analysis of biological thiols: determination of thiol components of disulfides and thioesters. Anal Biochem 154(1): 34-42.
- Getz, E. B., Xiao, M., Chakrabarty, T., Cooke, R. and Selvin, P. R. (1999). A comparison between the sulfhydryl reductants tris(2-carboxyethyl)phosphine and dithiothreitol for use in protein biochemistry. Anal Biochem 273(1): 73-80.
- Graham, D. E., Harich, K. C. and White, R. H. (2003). Reductive dehalogenation of monobromobimane by tris(2-carboxyethyl)phosphine. Anal Biochem 318(2): 325-328.
- Hashida, S. N., Miyagi, A., Nishiyama, M., Yoshida, K., Hisabori, T. and Kawai-Yamada, M. (2018). Ferredoxin/thioredoxin system plays an important role in the chloroplastic NADP status of Arabidopsis. Plant J 95(6): 947-960.
- Meyer, A. J., May, M. J. and Fricker, M. (2001). Quantitative in vivo measurement of glutathione in Arabidopsis cells. Plant J 27(1): 67-78.
- Newton, G. L., Dorian, R. and Fahey, R. C. (1981). Analysis of biological thiols: derivatization with monobromobimane and separation by reverse-phase high-performance liquid chromatography. Anal Biochem 114(2): 383-387.
- Riddles, P. W., Blakeley, R. L. and Zerner, B. (1983). Reassessment of Ellman's reagent. Methods Enzymol 91: 49-60.
- Tyagarajan, K., Pretzer, E. and Wiktorowicz, J. E. (2003). Thiol-reactive dyes for fluorescence labeling of proteomic samples. Electrophoresis 24(14): 2348-2358.
Article Information
Copyright
© 2019 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Hashida, S. and Kawai-Yamada, M. (2019). Detection of Disulfides in Protein Extracts of Arabidopsis thaliana Using Monobromobimane (mBB). Bio-protocol 9(5): e3183. DOI: 10.21769/BioProtoc.3183.
Category
Plant Science > Plant biochemistry > Protein > Modification
Biochemistry > Protein > Modification
Biochemistry > Other compound > Thiol
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









