- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Top Starch Plating Method for the Efficient Cultivation of Unicellular Red Alga Cyanidioschyzon merolae
Published: Vol 9, Iss 4, Feb 20, 2019 DOI: 10.21769/BioProtoc.3172 Views: 6505
Reviewed by: Dennis NürnbergMarion EisenhutMaksymilian Zienkiewicz

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Measuring Secretion of Capsidiol in Leaf Tissues of Nicotiana benthamiana
Teruhiko Kuroyanagi [...] Daigo Takemoto
Aug 5, 2018 6132 Views

Isolation of Powdery Mildew Haustoria from Infected Barley
Linhan Li [...] Pietro D. Spanu
Jul 20, 2019 7831 Views

Measurements of Root Colonized Bacteria Species
Vílchez Juan Ignacio [...] Huiming Zhang
Apr 5, 2021 6321 Views
Abstract
The unicellular red alga Cyanidioschyzon merolae has been used as a model photosynthetic eukaryote for various basic and applied studies, and several of these molecular genetics techniques have been reported. However, there are still improvements to be made concerning the plating method. The conventional plating method often generates diffuse colonies and single colonies cannot be easily isolated. To overcome these problems, we established a novel plating method for C. merolae, making use of melted cornstarch as the use of top agar plating in bacterial genetics. This method improved the formation of defined colonies in at least 4-fold higher efficiency than the conventional method, and made the handling procedure much easier than the previous method.
Keywords: Cyanidioschyzon merolaeBackground
Cyanidioschyzon merolae 10D is a unicellular red alga that inhabits sulfate acid hot springs (Matsuzaki et al., 2004). It has a simple cellular structure, composed of a minimum set of organelles: one nucleus, one mitochondrion, and one plastid in a cell. C. merolae is used as a model organism to investigate the basic architecture of eukaryotes. Thus, many molecular genetics techniques have been developed using this organism, including antisense suppression, transient expression, gene disruption, and stable insertion into the genome (Minoda et al., 2004; Ohnuma et al., 2008; Imamura et al., 2009; Imamura et al., 2010; Zienkiewicz et al., 2017a; Zienkiewicz et al., 2017b; Zienkiewicz et al., 2018). The plating method for the selection of transformants is important for transformation experiments. However, C. merolae shows considerably low plating efficiencies (less than 0.5%) on MA2 solid gellan gum plates. The direct spread of cells onto an MA2 solid gellan gum plate usually results in the death of the cells (Figure 1A). Hence, the plating method has been modified using the cornstarch embedding method which is commonly used for the cell-wall-less Chlamydomonas reinhardtii strain (Shimogawara et al., 1998). In the cornstarch embedding method, cells are grown on a starch bed prepared by spotting 20% slurry cornstarch on the MA2 solid gellan gum. Cells form colonies on the solid starch bed, but these are often diffused or blurred (Figure 1B).
To improve these points, we here devised a novel plating method using melted cornstarch (named in this protocol “top starch solution method”), similar to the top-agar plating method used in bacterial genetics. The 2% cornstarch in dH2O was melted at 98 °C for 10 min. The melted cornstarch solution (= top starch solution) was mixed with an equal volume of the liquid MA2 medium containing C. merolae cells, and poured onto the surface of the MA2 solid gellan gum. Cells formed defined colonies using this plating method (Figure 1C). The plating efficiency of the top starch solution method was more than 2.0%, while that of the conventional method was less than 0.5%. In the conventional method, it was necessary to prepare many starch beds on solid plates prior to use, but the top starch solution method only requires the pouring of the melted starch-cell mixture onto the solid medium, making the plating procedure quick and easy. In this report, we describe the detailed procedure of the top starch solution method.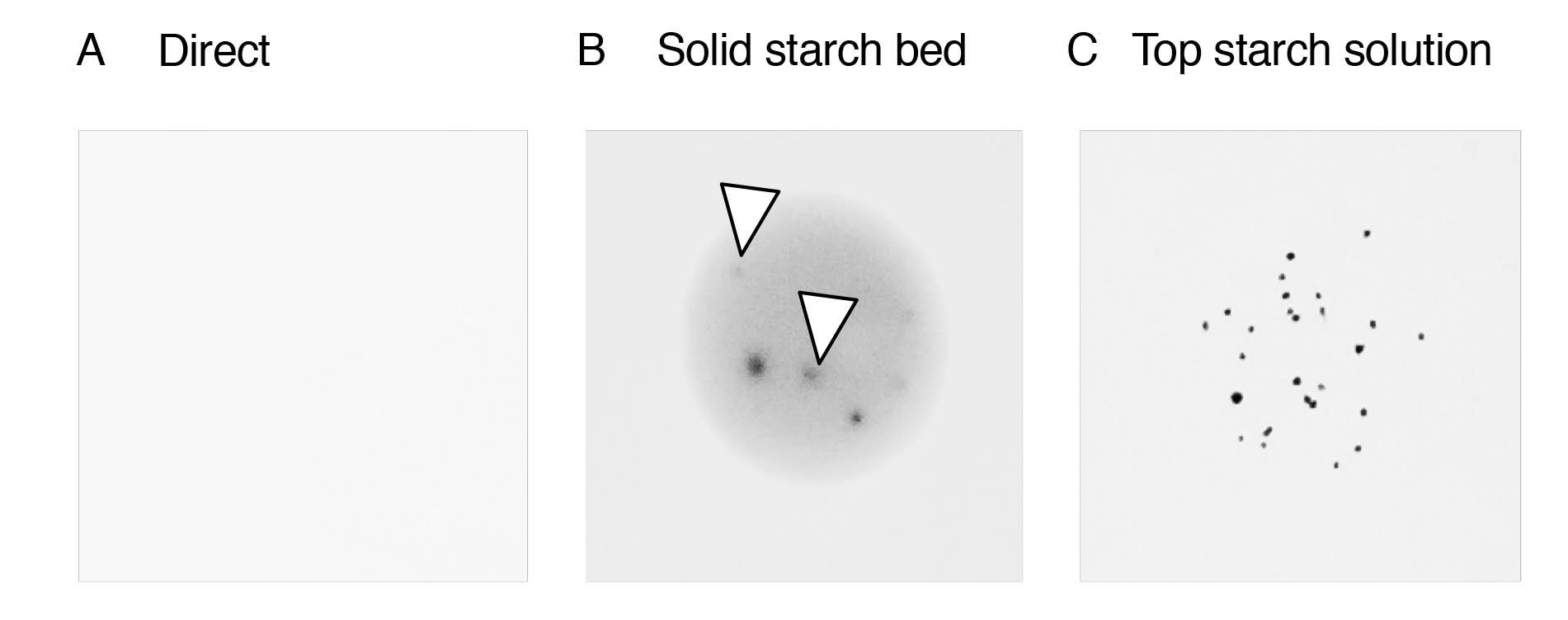
Figure 1. Effect of the top starch solution method on the colony formation in C. merolae. (A-C) In each approach, plates with the spotted cells were incubated for two weeks at 40 °C under continuous white light. A. Five hundred cells were directly spotted onto an MA2 solid gellan gum plate, which resulted in no visible colony formation. B. Solid starch beds were made using 15 μl of 20% slurry cornstarch in an MA2 liquid medium. Five hundred cells were spotted onto a solid cornstarch bed. Open triangles indicate diffused, blurred colonies. C. The 2% cornstarch in dH2O was melted at 98 °C for 10 min to make the top starch solution. The top starch solution was mixed with an equal volume of cells in MA2 medium. Fifteen microliter aliquots containing 500 cells were spotted onto the MA2 solid gellan gum plate.
Materials and Reagents
- Pipette tips (NIPPON Genetics Co., Ltd., catalog numbers: FG-102, FG-301, FG-401)
- Parafilm (LMS Co., Ltd., catalog number: PF, PM-996)
- 1.5 ml microtube (BM Equipment Co., Ltd., catalog number: NT-175)
- 2.0 ml microtube (WATSON Co., Ltd., catalog number: 132-620C)
- 50 ml centrifuge tube (Corning Incorporated., catalog number: 352196)
- AnaeroPouch (Mitsubishi Gas, catalog number: 2-3764-02)
- AnaeroPack (Mitsubishi Gas, catalog number: 2-3765-01)
- Sterilized plastic plate (Ø 90 mm) (As One Corporation., catalog number: GD90-15)
- Cyanidioschyzon merolae 10D [available as NIES-3377 from NIES collection, Tsukuba, Japan (http://mcc.nies.go.jp/index_en.html)]
- Cornstarch (Kawamitsu-Bussan, catalog number: 4901 486 02701 6)
- Gellan gum (FUJIFILM Wako Pure Chemical Corporation, catalog number: 073-03071)
- Uracil (Sigma-Aldrich, catalog number: U0750-5G)
- 5-Fluoroorotic acid monohydrate (5-FOA) (FUJIFILM Wako Pure Chemical Corporation, catalog number: 003234)
- Ethanol (FUJIFILM Wako Pure Chemical Corporation, catalog number: 055-00457)
- (NH4)2SO4 (FUJIFILM Wako Pure Chemical Corporation, catalog number: 019-03435)
- MgSO4∙7H2O (FUJIFILM Wako Pure Chemical Corporation, catalog number: 138-00415)
- H2SO4 (FUJIFILM Wako Pure Chemical Corporation, catalog number: 192-04696)
- H3BO3 (FUJIFILM Wako Pure Chemical Corporation, catalog number: 021-02195)
- MnCl2∙4H2O (FUJIFILM Wako Pure Chemical Corporation, catalog number: 133-00725)
- ZnCl2 (FUJIFILM Wako Pure Chemical Corporation, catalog number: 268-01022)
- Na2MoO4∙2H2O (FUJIFILM Wako Pure Chemical Corporation, catalog number: 198-02471)
- CoCl2∙6H2O (FUJIFILM Wako Pure Chemical Corporation, catalog number: 038-03681)
- CuCl2∙2H2O (FUJIFILM Wako Pure Chemical Corporation, catalog number: 039-04135)
- KH2PO4 (FUJIFILM Wako Pure Chemical Corporation, catalog number: 169-04245)
- CaCl2∙2H2O (FUJIFILM Wako Pure Chemical Corporation, catalog number: 038-19735)
- FeCl3∙6H2O (FUJIFILM Wako Pure Chemical Corporation, catalog number: 090-02802)
- Na2EDTA (FUJIFILM Wako Pure Chemical Corporation, catalog number: 345-01865)
- 20% slurry cornstarch stock solution (see Recipes)
- MA2 medium (Kobayashi et al., 2010) (see Recipes)
- MA2 solution I
- A6 minor salts
- MA2 solution II
- MA2 solution III
- MA2 solution IV
- MA2 solid gellan gum plate (see Recipes)
Equipment
- Pipettes (Gilson, Inc., catalog numbers: F123600, F123601, F123602)
- Refrigerated centrifuge (Koki Holdings Co., Ltd., model: CF16RXII)
- Microcentrifuge (TOMY Seiko Co., Ltd., model: MX150)
- 500 ml flask (IWAI, catalog number: 4442FK500)
- Heat block (Chiyoda Science Co., Ltd., model: MiniT-100)
- Bio incubator (TOMY Seiko Co., Ltd., model: CLE-303)
- Clean bench (Panasonic Healthcare Co., Ltd., model: MCV-131BNF)
- Vortex mixer (M & S Instruments Inc., model: SI-0286)
- Sterile filter (Ø 0.22 μm, Merck, catalog number: SLGV033RS)
Procedure
- Prepare the MA2 solid gellan gum plate and the 20% slurry cornstarch stock solution (see Recipes).
- Mix 100 μl of the cornstarch slurry stock solution and 900 μl of sterile dH2O in a 2 ml microtube.
- Centrifuge the microtubes at 500 x g for 1 min at room temperature and discard the supernatant.
- Add 900 μl of sterile dH2O and mix vigorously.
- Centrifuge again at 500 x g for 1 min at room temperature and discard the supernatant.
- Add 900 μl of sterile dH2O and mix vigorously.
- Incubate the microtube at 98 °C for 10 min but continue to mix thoroughly by inverting the tube every 2 min.
- Cool down at room temperature for 10 min (= top starch solution).
Note: Steps 8-11 should be performed in a laminar flow cabinet. - Add 1 ml of MA2 culture medium containing C. merolae cells (in case of wild type, 500 cells by microscopy observation) to the top starch solution and mix gently.
- Pour the mixture onto the surface of the MA2 solid gellan gum plate and spread the mixture over the whole plate area by tilting the plate to spread the top starch solution uniformly.
- Leave the plate open for 15-20 min to allow the top starch solution to solidify. After this step, the surface of the top starch is still wet, and the plate should be incubated with the top starch solution pointing upwards for the entire time.
- Incubate the MA2 solid gellan gum plate at 40 °C under continuous white light (35-50 μmol m-2 s-1) supplemented with 5% CO2 in AnaeroPack with AnaeroPouch.
- After 2-4 weeks, colonies of the transformed cells appear on the MA2 solid gellan gum plate.
Recipes
- 20% slurry cornstarch stock solution (Fujiwara and Ohnuma, 2018)
- Add 10 g of cornstarch and 40 ml of dH2O to the 50 ml conical tube
- Mix the tube well using a vortex mixer
- Centrifuge the tube at 1,200 x g for 5 min at 4 °C
- Discard the supernatant by decantation
- Resuspend the pellet in 40 ml of 100% of ethanol using a vortex mixer
- Centrifuge the tube at 1,200 x g for 5 min at 4 °C
- Discard the supernatant by decantation
- Fill up to 50 ml with 75% of ethanol
- Store the tube at 4 °C, wrapped using parafilm until use
- MA2 medium
- Mix 100 ml of MA2 solution I, 10 ml of MA2 solution II, 1 ml of MA2 solution III and 885 ml of dH2O in a glass bottle and sterilize by autoclaving
- Add 4 ml of MA2 solution IV to the mixture in a laminar flow cabinet
MA2 solution I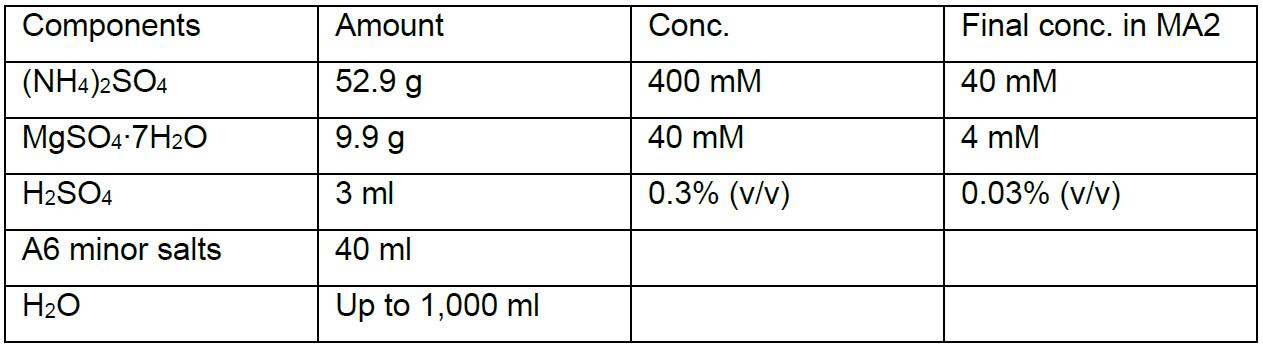
A6 minor salts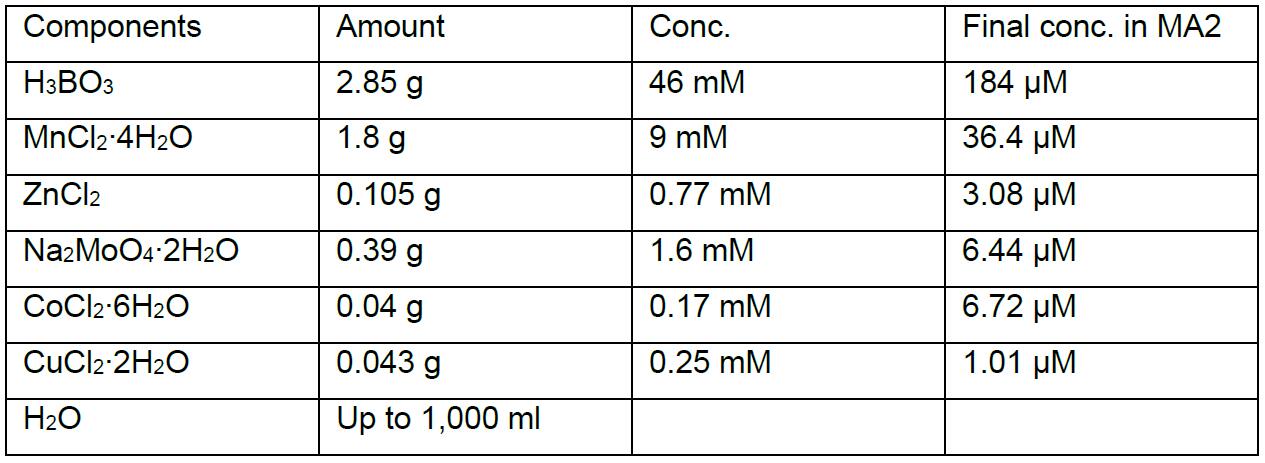
MA2 solution II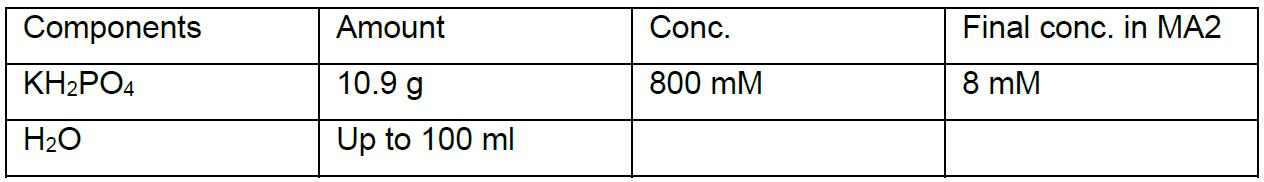
MA2 solution III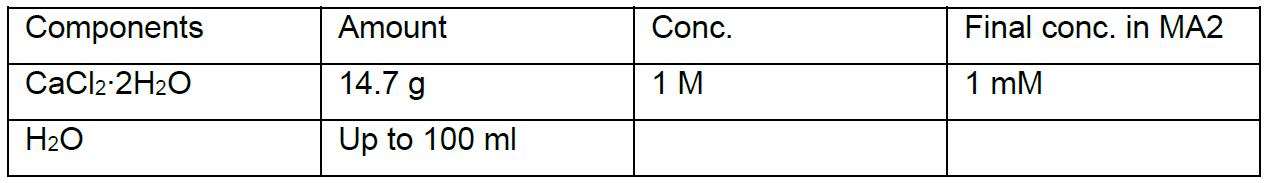
MA2 solution IV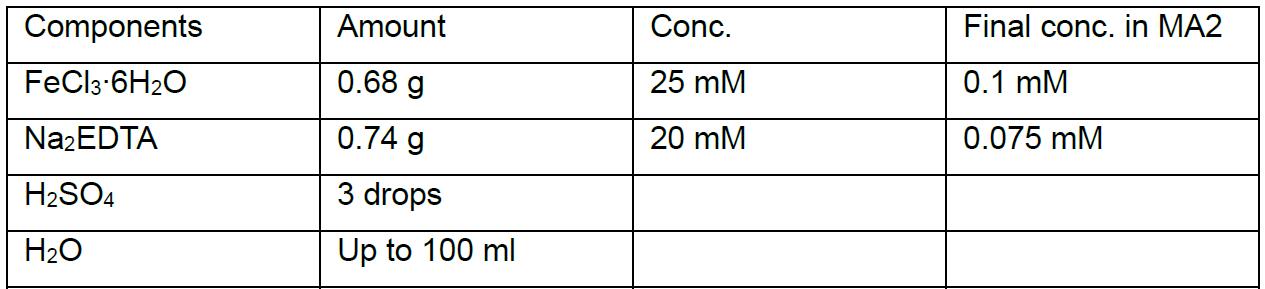
Note: This MA2 solution IV should be sterilized by filtration and stored at 4 °C in the dark. - MA2 solid gellan gum plate (for making 9-10 plates)
- Mix 30 ml of MA2 solution I, 3 ml of MA2 solution II, 300 μl of MA2 solution III, and 70 ml of dH2O in a 500 ml flask. Mix 200 ml of dH2O and 1.5 g of gellan gum in another 500 ml flask. Sterilize these flasks by autoclaving, and subsequent plate preparation should be performed in a laminar flow cabinet
- After autoclaving, mix both solutions and add 1.2 ml of MA2 solution IV. In case of 5-FOA selection to isolate the URA deficient mutants (for example applying URA marker recycling system, Takemura et al., 2018; Takemura et al., 2019), add directly 150 mg of uracil (final concentration: 0.5 mg/ml) and 240 mg of 5-FOA (final concentration: 0.8 mg/ml) in powder to the flask
- Pour the mixed solution into the sterilized plastic plates
- Dry the plates at room temperature for 15 min
- Store at 4 °C in the dark
Acknowledgments
This protocol was adapted from Takemura et al. (2018). The authors thank Y. Ide for technical assistance. This study was supported by MEXT/JSPS KAKENHI (Grant numbers: 17K07438 to S.I., 17K07439 to Y.K.) and by Advanced Low Carbon Technology Research and Development Program (ALCA) of Japan Science and Technology Agency (JST) to K.T.
Competing interests
No competing interests declared.
References
- Fujiwara, T. and Ohnuma, M. (2018). Procedures for transformation and their applications in Cyanidioschyzon merolae. In: Kuroiwa, T., Miyagishima, S.Y., Matsunaga, S., Sato, N., Nozaki, H., Tanaka, K. and Misumi, O. (Eds.). Cyanidioschyzon merolae. Singapore, Springer Singapore: 87-103.
- Imamura, S., Kanesaki, Y., Ohnuma, M., Inouye, T., Sekine, Y., Fujiwara, T., Kuroiwa, T. and Tanaka, K. (2009). R2R3-type MYB transcription factor, CmMYB1, is a central nitrogen assimilation regulator in Cyanidioschyzon merolae. Proc Natl Acad Sci U S A 106(30): 12548-12553.
- Imamura, S., Terashita, M., Ohnuma, M., Maruyama, S., Minoda, A., Weber, A. P., Inouye, T., Sekine, Y., Fujita, Y., Omata, T. and Tanaka, K. (2010). Nitrate assimilatory genes and their transcriptional regulation in a unicellular red alga Cyanidioschyzon merolae: genetic evidence for nitrite reduction by a sulfite reductase-like enzyme. Plant Cell Physiol 51(5): 707-717.
- Kobayashi, Y., Ohnuma, M., Kuroiwa, T., Tanaka, K., and Hanaoka, M. (2010). The basics of cultivation and molecular genetic analysis of the unicellular red alga Cyanidioschyzon merolae. Endocytobiosis Cell Res 20: 53-61.
- Matsuzaki, M., Misumi, O., Shin-I, T., Maruyama, S., Takahara, M., Miyagishima, S. Y., Mori, T., Nishida, K., Yagisawa, F., Nishida, K., et al. (2004). Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature 428(6983): 653-657.
- Minoda, A., Sakagami, R., Yagisawa, F., Kuroiwa, T. and Tanaka, K. (2004). Improvement of culture conditions and evidence for nuclear transformation by homologous recombination in a red alga, Cyanidioschyzon merolae 10D. Plant Cell Physiol 45(6): 667-671.
- Ohnuma, M., Yokoyama, T., Inouye, T., Sekine, Y. and Tanaka, K. (2008). Polyethylene glycol (PEG)-mediated transient gene expression in a red alga, Cyanidioschyzon merolae 10D. Plant Cell Physiol 49(1): 117-120.
- Shimogawara, K., Fujiwara, S., Grossman, A. and Usuda, H. (1998). High-efficiency transformation of Chlamydomonas reinhardtii by electroporation. Genetics 148(4): 1821-1828.
- Takemura, T., Imamura, S., Kobayashi, Y. and Tanaka, K. (2018). Construction of a selectable marker recycling system and the use in epitope tagging of multiple nuclear genes in the unicellular red alga Cyanidioschyzon merolae. Plant Cell Physiol 59(11): 2308-2316.
- Takemura, T., Imamura, S., Kobayashi, Y. and Tanaka, K. (2019). Preparation of introducing DNA fragment for multiple modification of chromosomal loci in the unicellular red alga Cyanidioschyzon merolae. Bio-protocol (In-review).
- Zienkiewicz, M., Krupnik, T., Drożak, A., Golke, A. and Romanowska, E. (2017a). Chloramphenicol acetyltransferase-a new selectable marker in stable nuclear transformation of the red alga Cyanidioschyzon merolae. Protoplasma 254(1): 587-596.
- Zienkiewicz, M., Krupnik, T., Drożak, A., Golke, A. and Romanowska, E. (2017b). Transformation of the Cyanidioschyzon merolae chloroplast genome: prospects for understanding chloroplast function in extreme environments. Plant Mol Biol 93(1-2): 171-183.
- Zienkiewicz, M., Krupnik, T., Drożak, A., Wasilewska, W., Golke, A. and Romanowska, E. (2018). Deletion of psbQ' gene in Cyanidioschyzon merolae reveals the function of extrinsic PsbQ' in PSII. Plant Mol Biol 96(1-2): 135-149.
Article Information
Copyright
© 2019 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Takemura, T., Kobayashi, Y., Imamura, S. and Tanaka, K. (2019). Top Starch Plating Method for the Efficient Cultivation of Unicellular Red Alga Cyanidioschyzon merolae. Bio-protocol 9(4): e3172. DOI: 10.21769/BioProtoc.3172.
Category
Plant Science > Phycology > Cell isolation and culture
Cell Biology > Cell isolation and culture > Cell isolation
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










